SATURATION MUTAGENESIS OF L177 IN HALOALKANE DEHALOGENASE LINB
Radka Chaloupková1, Jana Sýkorová1, Zbyněk Prokop1, Andrea Jesenská1, Marta Monincová1, Martina Pavlová1, Yuji Nagata2 and Jiří Damborský1
1National Centre for Biomolecular Research, Masaryk University, Kotlářská 2, 611 37 Brno, Czech Republic
2Graduate School of Life Sciences, Tohoku University, Katahira, Sendai 980-8577, Japan
Enzyme LinB is the haloalkane dehalogenase from bacterium Sphingomonas paucimobilis UT26. It is involved in a biochemical pathway for degradation g-hexachlorocyclohexane. LinB catalyses hydrolytic dehalogenation of broad range of halogenated aliphatic compounds [1]. The amino acid in position 177 was identified as a very important determinant of catalytic properties of LinB by structural analysis [2] and by comparison of its protein sequence with other family members: (i) L177 is positioned in the mouth of the entrance tunnel leading the enzyme active site and is pointing directly to the tunnel and (ii) L177 is the most variable residue of the active site pocket among different haloalkane dehalogenases. L177 of the wild type enzyme was therefore replaced by every other amino acid and then the effect of mutations on enzyme activity was studied. Construction of the protein variants was conducted in two successive rounds. In the first round, L177 was replaced by A, C, G, F, K, T and W, respectively. The specific activities of the first set of mutants were statistically analysed but results from this analysis were not statistically significant. For that reason the first set of mutants was complemented with the second set of mutants comprising replacement of L177 by D, E, H, I, M, N, P, Q, R, S, V and Y, respectively.
All seven protein variants of the first set could be overexpressed in Escherichia coli and showed activity with at least some of the substrates used for characterization. In the second set, two out of twelve protein variants (L177E and L177N) could not be overexpressed in E. coli, while other two variants (L177P and L177I) did not show activity with any of the substrates. Circular dichroism spectra were recorded for all proteins purified in the second set and two inactive mutants showed spectra different from wild-type LinB and other mutants (Fig. 1), suggesting a decrease in the number of amino acids in a-helical conformation and protein unfolding.
Successfully purified enzymes from both sets were kinetically characterized using a gas chromatography. Compounds 1-chlorobutane and 1,2-dibromoethane were selected as the substrates for steady-state kinetic measurements because they often serve as the reference compounds for characterization of the haloalkane dehalogenases. Dehalogenation of 1-chlorobutane showed typical Michaelis-Menten dependence, while dehalogenation of 1,2-dibromoethane showed substrate inhibition. Furthermore, the specific activities of all prepared enzymes were determined for twelve different substrates (1-chlorobutane, 1-chlorohexane, 1-bromobutane, 1-iodobutane, 1,2-dichloroethane, 1,2-dibromoethane, 1,3-diidopropane, 1,2-dichloropropane, 1,2,3-trichloropropane, chlorocyclohexane, bromocyclohexane and 3-chloro-2-methylpropene) and statistically analysed by Principal Component Analysis. The first and only important component explained 45.8% of the data variance. Catalytic activity of mutant protein correlated mainly with the size of amino acid introduced to the position 177.
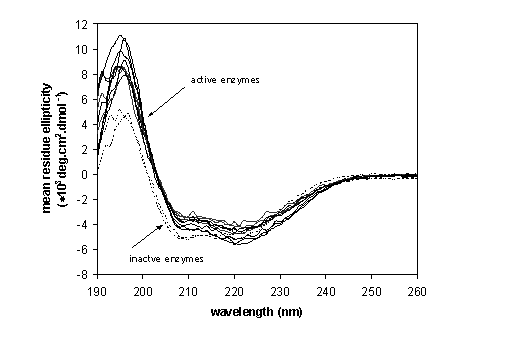
Fig. 1. Far-UV circular dichroism spectra of the wild-type haloalkane dehalogenase LinB, L177D, L177H, L177M, L177Q, L177R, L177S, L177V, L177Y mutants (solid lines) and L177P, L177I mutants (dashes lines). The spectra were measured at room temperature with the protein concentration 0.3 mg/ml in 50 mM phosphate buffer (pH 7.5) using the JASCO J-810 spectropolarimeter.
References:
1. Y. Nagata, K. Miyauchi, J. Damborský, K. Manová, A. Ansorgová & M. Takagi, Appl. Env. Microbiol., 63 (1997) 3707-3010.
2. J. Marek, J.
Vévodová, I. Kutá-Smatanová, Y. Nagata, L. A. Svensson, J. Newman, M. Takagi & J. Damborský, Biochemistry,
39 (2000) 14082-14086.