Molecular structure as a prerequisite for function – Laboratory of Structure and Function of Biomolecules of IBT
J. Dohnálek, T. Skálová, J. Dušková, P. Kolenko, K. Adámková, M. Trundová, L. Švecová, T. Kovaľová, M. Malý, J. Hrubý, B. Husťáková, J. Stránský, J. Hašek, T. Kovaľ
Institute of Biotechnology of the Czech Academy of Sciences, Průmyslová 595, 25250 Vestec, Czech Republic
dohnalek@ibt.cas.cz
Our understanding of function of a biological macromolecule requires information about its primary, secondary, tertiary, and quaternary structure. Targeted manipulation of structural features then enables direct observation of functional consequences. The complete understanding of the structure-function relationship in biomolecules would provide us with a full view of the rules governing biological molecular systems and enable relatively easy control of their properties. Typically, this complete knowledge is never achieved as we are dealing with multivariable systems with incomplete description. However, we take the advantage of availability of a number of structural and biophysical experimental techniques, together with functional assays of the studied systems to form a picture which is as complete as possible and also precise and accurate.
Our structural analysis methods include single crystal X-ray crystallography, small-angle X-ray scattering (SAXS), and cryogenic electron microscopy (cryoEM). Crystallographic analysis of proteins, protein:protein, protein:ligand, or protein:nucleic acids complexes provides structural information at high or even atomic resolution, uncovering key interaction details, new types of three-dimensional protein structure, new types of bonds, intrinsic structural properties, identification of ligands or quantification of metal clusters occupancy. Application of SAXS results in information on low-resolution arrangement, protein oligomerization or induced structural changes. CryoEM single particle imaging is used to elucidate structure of difficult-to-crystallize multi-protein complexes.
Apart from structural analysis we utilize a range of methods of molecular biophysics. They provide priceless data on protein or complex stability (Differential Scanning Fluorimetry), structural state (Circular Dichroism), intermolecular interactions (Microscale Thermophoresis, Isothermal Calorimetry), size and aggregation/oligomerization of molecules (Dynamic Light Scattering, SAXS) or complex formation/size distribution (Mass Photometry).
In our studies of bilirubin oxidase we have uncovered the nature and role of a new covalent bond formed between two amino acids of the enzyme (Fig. 1) [1]. The structure-function study of the first representative of GH151 family of α-l-fucosidases revealed not only an unexpected oligomerization pattern but also active site complementation [2]. Formation of an unprecedented complex between protein HelD and mycobacterial RNA polymerase helped explain HelD function and its emerging role in antibiotic resistance [3-4]. Our atomic-resolution study of S1 nuclease complexes with products of RNA cleavage provided insights into the intimate details of intermolecular interactions including dynamics of substrate/product binding [5]. The principle characteristics of protein:protein interactions between immune cell surface receptor NKR-P1 and its cognate ligand LLT1 pointing towards formation of larger zipper-like structures were uncovered in a recent collaborative study [6].
Mainly the structural analysis aspects of our studies will be presented to illustrate the power of this approach in explanation of functionality relevant for biomedical and biotechnological applications. Some of our contributions to the development of methods of structural analysis will be also mentioned [7-8].
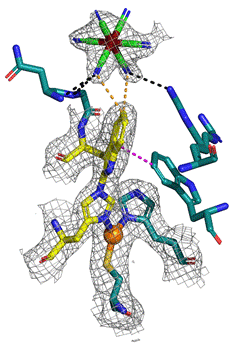
Figure 1. Detail of a crystal structure showing an unusual covalent bond between Trp and His in the surface active site of bilirubin oxidase from Myrothecium verrucaria, PDB ID 3I3J [1]. Coordination of T1 copper is represented as covalent bond. Colour coding: T1 site and oxidation site residues – carbon teal; covalent adduct Trp-His – carbon yellow; copper orange, iron brown; ferricyanide – carbon green. Interactions of [Fe(CN)6]3- - black dashed line, in yellow VDW interaction with Trp396. Magenta dashed line – CH-p interaction.
1. T. Kovaľ, L. Švecová, L. H. Østergaard, T. Skalova, J. Dušková, J. Hašek, P. Kolenko, K. Fejfarová, J. Stránský, M. Trundová, J. Dohnálek J. Sci. Rep., 9, (2019), 13700.
2. T. Koval'ová, T. Kovaľ, J. Stránský, P. Kolenko, J. Dušková, L. Švecová, P. Vodičková, V. Spiwok, E. Benešová, P. Lipovová, J. Dohnálek. FEBS J., 289, (2022), 4998.
3. T. Kouba, T. Koval', P. Sudzinová, J. Pospíšil, B. Brezovská, J. Hnilicová, H. Šanderová, M. Janoušková, M. Šiková, P. Halada, M. Sýkora, I. Barvík, J. Nováček, M. Trundová, J. Dušková, T. Skálová, U. Chon, K. S. Murakami, J. Dohnálek, L. Krásný. Nat. Commun., 11, (2020), 6419.
4. P. Sudzinová, H. Šanderová, T. Kovaľ, T. Skálová, N. Borah, J. Hnilicová, T. Kouba, J. Dohnálek, L. Krásný. FEMS Microbiol. Rev., (2022), doi: 10.1093/femsre/fuac051. Epub ahead of print.
5. K. Adámková, T. Koval', L. H. Østergaard, J. Dušková, M. Malý, L. Švecová, T. Skálová, P. Kolenko, J. Dohnálek. Acta Crystallogr. D Struct. Biol., 78, (2022), 1194.
6. J. Bláha, T. Skálová, B. Kalousková, O. Skořepa, D. Cmunt, V. Grobárová, S. Pazicky, E. Poláchová, C. Abreu, J. Stránský, T. Kovaľ, J. Dušková, Y. Zhao, K. Harlos, J. Hašek, J. Dohnálek, O. Vaněk. Nat. Commun., 13, (2022), 5022.
7. M. Malý, K. Diederichs, J. Dohnálek, P. Kolenko. IUCrJ., 7, (2020), 681.
8. P. Kolenko, J. Stransky, T. Koval', M. Maly, J. Dohnalek. J. Appl. Cryst., 54, (2021), 996.
This work was supported by MEYS (LM2018127, LM2023042), CSF (23-06295S, 18-10687S, 20-12109S), ERDF (CZ.02.1.01/0.0/0.0/16_013/0001776, CZ.02.1.01/0.0/0.0/15_003/0000447) and AS CR (86652036).