Herpesviral HCMV UL141 antagonist development to block TRAIL death receptor binding
I. Nemčovičová1, J. Kóňa2, M. Poláková2, T. Klunda2, A. Bitala1, M. Benko1, M. Nemčovič2
1Biomedical Research Center, Slovak Academy of Sciences, Bratislava, Slovakia
2Institute of Chemistry, Slovak Academy of Sciences, Bratislava, Slovakia
viruivka@savba.sk
Numerous studies have demonstrated that human cytomegalovirus (HCMV) encodes countermeasures against a spectrum of immune responses [1-5]. This arsenal of immunomodulatory functions is likely a reflection of the natural history of the virus, providing the capacity to establish lifelong infections of the host as well as to reinfect people with an existing infection despite the presence of a substantial immune response. The complexity of these immunological interactions is being studied extensively. HCMV has developed many sophisticated mechanisms targeting host immunity and has become a paradigm for viral immune evasion. Suffice it to say that HCMV-encoded gene functions target antigen presentation by major histocompatibility complex (MHC) class I and class II molecules, utilize cytokine mimicry to exert paracrine functions against immune cells and encode proteins that antagonize the range of innate immune responses directed against the virus. We have been studying several of viral immune modulatory genes over the last decade, primarily focusing on those that target and/or intersect with Ig-like- and TNF-family signaling [2-5]. The fact that ablating a single strategy can markedly impact infection suggests that multiple Achilles’ heals may be exploitable for antiviral agents or biotherapeutics development.
HCMV commensurately uses own proteins to restrict NK activating ligands and receptors on the surface of infected cells. An extra-ordinarily effective strategy is encoded by the HCMV UL141 protein, which inhibits surface expression of the NK activating ligands CD155 and CD112 [5] and also binds death receptors for the TNF apoptotic ligand TRAIL [2,4]. This pleiotropy of UL141 is required for its broad and potent inhibition of NK cells, and a viral mutant lacking UL141 is highly susceptible to NK killing, revealing how disrupting the function of a single HCMV gene can tip the balance in favor of host defense.
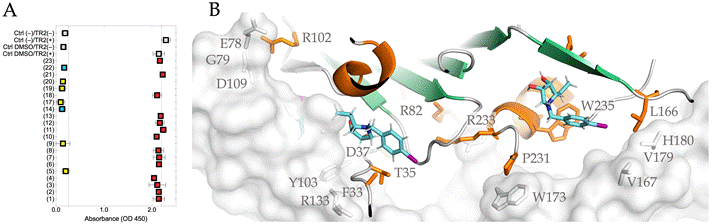
Figure 1. ELISA-like TMB assay (A), where dynabeads are coated by HCMV UL141, has revealed seven compounds that blocks TRAIL-R2 receptor binding (OD450 <2.0) (yellow and cyan squares). SPR binding assay revealed two compounds (14) and (22) that bind HCMV UL141 with higher affinity. Both compounds (in cyan sticks) were docked to HCMV UL141 structure to reveal specific binding site (B), while TRAIL-R2 receptor is also shown (in grey surface).
In this work, we sought to develop the short peptide or synthetic compound (UL141 antagonist) based on our recent crystal structure that would specifically binds viral UL141 to block receptor binding thus prevent the viral action. This is relevant, as the UL141 is the most abundant HCMV protein on plasma membrane and it is also a component of the virion. Within first part, we sought to test a small library of synthetized compounds (potential UL141 antagonists) that would block the receptor binding in vitro, on the cell, or virion surface. Series of compounds that have been tested are of glycomimetics structures consisting of various saccharide units linked with non-saccharide. In particular; non-ionic glycolipids, ‘click’-conjugates or iminosugars. The ELISA-like TMB assay has been used in combination with dynabeadsTM coating to test whether the compound could block the TRAIL-R2 binding. The most promising compounds are (14) and (22) (out of 23 tested) that have proven the ability to block UL141/TRAIL-R2 complex formation. SPR kinetics analysis was used to determine the binding constants (KD). The affinities to UL141 were determined to 24 mM and 29 mM for compound 14 and 22, respectively. Moreover, both compounds were docked to UL141 structure to reveal specific binding sites. The successful compounds will be further optimized by using in silico methods to target particular epitope on viral glycoprotein UL141 derived from our structural analysis.
1. Picarda, G. & Benedict, C. A. J. Immunol., 200, (2018), 3881.
2. Nemčovičová, I., Benedict, C. A. & Zajonc, D. M. PLoS Pathog., 9, (2013), 1003224.
3. Nemčovičová, I. & Zajonc, D. M. Acta Crystallogr. D Biol. Crystallogr., 70, (2014), 851.
4. Smith, W., Tomasec, P., Aicheler, R., Loewendorf, A. Nemčovičová, I. et al. Cell Host Microbe., 13, (2013), 324.
5. Tomašec, P., Wang, E. C. Y., Davison, A. J., Vojtešek, B., Armstrong, M. et al. Nat. Immunol., 6, (2005), 181.
This research was funded by the contribution of the Slovak Research and Development Agency under the project APVV-19-0376; and the contribution of the Scientific Grant Agency of the Slovak Republic under the grant VEGA-02/0026/22.