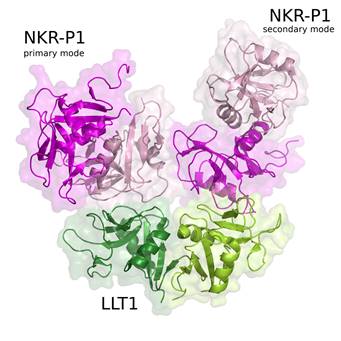
Figure 1. The crystal structure (PDB code 5MGT) revealed two binding modes of immune receptor NKR-P1 with its protein ligand LLT1, here denoted as primary and secondary binding mode.
1 Institute of Biotechnology, Czech Academy of Sciences, BIOCEV, Průmyslová 595, 252 50 Vestec, Czech Republic
Natural killer cells are white blood cells able to kill tumour, virus-infected or stressed cells. NKR-P1 is an immune receptor on the surface of human natural killer cells and LLT1 protein is its binding partner on the surface of another cell interacting with the NK cell. Both NKR-P1 and LLT1 have extracellular domains of the same C-type lectin-like (CTL) fold. LLT1 is expressed on activated monocytes, B cells and cancer cells. Therefore, structural knowledge of the interaction between NKR-P1 and LLT1 is important for understanding physiological and pathogenic processes in the immune system.
Crystal structure of human NKR-P1 in complex with LLT1 was determined using diffraction data collected at the Diamond Light Source (Harwell, UK) at beamline I03 with resolution 1.9 Å [1].


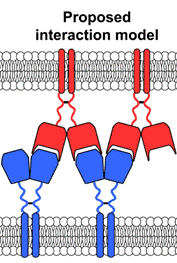
Besides this structure, we solved also three structures of LLT1 (monomeric, dimeric and dimeric fully glycosylated, [2]) and two structures of NKR-P1 (glycosylated and deglycosylated dimer, [1]). These structures confirm that NKR-P1 and LLT1 systematically differ in the way of their dimerization. The structure of the NKR-P1:LLT1 complex showed that different mode of dimerization of the binding partners is important for their geometrical arrangement. Moreover, the structure of the complex revealed two different binding modes between the proteins (Fig. 1, primary and secondary binding mode) and virtually “infinite” chains of both proteins existing in the crystal. Biological relevance of both binding modes was confirmed by mutational analysis and clustering of the receptors with ligands was observed also on cell surface using super-resolution microscopy. The proposed interaction model is shown in Figure 2.
This study was supported by the Czech Science Foundation grant 18-10687S, the Ministry of Education, Youth and Sports of the Czech Republic grant LTC17065 (in the frame of the COST Action CA15126 MOBIEU), and the European Regional Development Fund (CZ.02.1.01/0.0/0.0/15_003/0000447). Computational resources were supplied by the project Infrastruktura CZ (e-INFRA LM2018140) provided within the program Projects of Large Research, Development, and Innovations Infrastructures. We acknowledge using CF Biophysical methods of CMS, CIISB, Instruct-CZ Centre, supported by MEYS CR (LM2018127).
1. J. Bláha, T. Skálová, B. Kalousková, O. Skořepa, D. Cmunt, V. Grobárová, S. Pazicky, E. Poláchová, C. Abreu, J. Stránský, T. Kovaľ, J. Dušková, Y. Zhao, K. Harlos, J. Hašek, J. Dohnálek and O. Vaněk, Nat. Comm. 13, (2022), 5022.
2. T. Skálová, J. Bláha, K. Harlos, J. Dušková, T. Kovaľ, J. Stránský, J. Hašek, O. Vaněk and J. Dohnálek, Acta Crystallogr. D71, (2015), 578-591.