Structural studies of Si3 endolysin mutants
D. Peramotava1, P. Grinkevich1, T. Prudnikova1, I. Kutá Smatanová1, Daria. V. Vasina2
1University of South Bohemia in České Budějovice, Branišovská 1760, 370 05 České Budějovice, Czech Republic
2Laboratory of Pathogen Population Variability Mechanisms, N. F. Gamaleya National Research Center for Epidemiology and Microbiology, 18 Gamaleya St., 123098 Moscow, Russia
daschaperemotova@gmail.com
For more than half a century, antibiotics have been used as the most common tool in the fight against infectious diseases. However, due to the misuse and overuse of these substances, an increasing number of bacteria are emerging that are completely resistant to all currently known antibiotics (multidrug resistant). The emergence of these types of bacteria renders most, if not all, available antibiotics ineffective. This situation is a global health crisis that threatens effective prevention and control of ever-increasing infectious diseases. This crisis is also exacerbated by increasingly sophisticated development new antibiotics.
A way out of this crisis may be the use of novel substances in the fight against infectious diseases. One of the potential candidates are endolysins. These enzymes, originally derived from bacteriophages, are peptidoglycan hydrolases that destroy bacterial cell wall. They have high specificity, much higher capability and efficiency, and also have not been found to develop resistance [1, 2]. An extremely fast and efficient lysis of peptidoglycan leads to a sudden drop in turgor pressure and osmotic lysis, causing bacterial cell death [3]. The most important aspect for us is how molecular engineering can be used to change lytic spectrum of endolysins [4].
The aim of our work was to obtain crystals of three Si3 endolysin mutants (lysAAA, lysHE and lysECD7-SMAP), followed by the study of their structure.
In the course of the work carried out, we obtained crystals of high diffraction quality for all three mutants presented above, which allowed us to study their structure in sufficient detail. X-ray diffraction data wore collected at BESSY II, Helmholtz Zentrum Berlin and processed used XDS-app and CCP4 software. The main statistical parameters are given in Table 1.
Table 1. Selected data collection statistics
|
Parameters |
lysECD7-SMAP |
lysAAA |
LysHE |
|
Resolution (Å) |
1,7 |
1.43 |
2 |
|
Average unit cell (a, b, c, Å; α=β=γ (℃)) |
48.66, 58.03, 196.63, 90.00 |
66.307, 76.548, 65.633 90.00 |
43.183, 66.381, 119.26 90.00 |
|
Space group |
P 21 21 21 |
C 2 2 21 |
P 2 21 21 |
|
Number of molecules in asymmetric unit |
4 |
1 |
2 |
|
R |
0.216 |
0.166 |
0.183 |
|
Rfree |
0.260 |
0.204 |
0.221 |
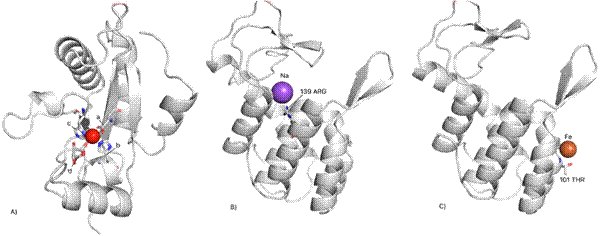
Figure 1. Secondary structures of mutants. A) the lysECD7-SMAP mutant molecule has a Zn atom (red ball) that is stabilized by amino acids 62 HIS (a), 69 ASP (b) and 117 HIS (c). Also shown is a glycerol molecule (d) located in the region of the Zn atom; C) lysAAA mutant with Na ion (purple ball); C) lysHE mutant, the structure contains the Fe ion (orange ball).
As can be seen from Figure 1, the structure of the lysECD7-SMAP mutant is fundamentally different from the structures of the lysAAA and lysHE mutants, which, in turn, are similar to each other. It is worth noting separately the presence of different metal atoms in the structure of each molecule; however, only in the lysECD7-SMAP mutant is the Zn atom stabilized by amino acid residues. ECD7-SMAP mutant has a 19-amino-acid C-terminal tail which changes overall conformation that may impact its enzymatic activity. It is, therefore, our main focus of further structural studies.
1. López, R.; García, E.; García, P., Drug Discovery Today: Therapeutic Strategies, 1, 2004, pp 469–474;
2. Fischetti, V.A., Trends Microbiology, 13, 2005, pp 491–496;
3. Vollmer, W.; Biochimica et Biophysica Acta (BBA) – Biomembranes, 1778, 2008, pp 1714–1734;
4. Kashani, H.H., Schmelcher, M., Sanzalipoor, H., Hosseini, E.S.Moniri, R., Clin. Microbiol. Rev., 31, 2018, e00071-17