Zn2+ to Ni2+ Exchange in Zn-Dependent S1 Nuclease
J. Hrubý1,2, P. Kolenko1,2, K. Adámková2,3, B. Husťáková2,3, M. Malý1,2, L. H. Østergaard4, T. Kovaľ2, J. Dohnálek2
1Czech Technical University in Prague, Břehová 7, 115 19 Prague, Czech Republic
2Institute of Biotechnology of the Czech Academy of Sciences, Biocev, Průmyslová 595, Vestec,
3University of Chemical and Technology Prague, Technická 5, Prague, Czech Republic
4Dept. of Agile Protein Screening, Novozymes A/S, Krogshoejvej 36, Bagsvaerd, Denmark
hrubyj18@fjfi.cvut.cz
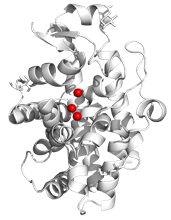
The single-strand-specific S1 nuclease from Aspergillus oryzae is a metalloenzyme with a widespread use for biochemical analysis of nucleic acids [1,2]. It is a globular protein with a secondary structure composed mainly of a-helices (Fig. 1). Its activity depends on the presence of three Zn2+ ions in the active site: Two Zn2+ ions of the cluster are buried at the bottom of the active site, while the third Zn2+ ion is closer to the surface of the nuclease. The core of the active site is composed of nine residues coordinating the zinc cluster.
We studied the possibility of replacing Zn2+ with Ni2+ using the X-ray anomalous dispersion and other biophysical assays. The mixture of deglycosylated S1 nuclease, chelating agent ethylenediaminetetraacetic acid and NiCl2 in a molar ratio of 1: 5: 10, respectively, was crystallized using the vapor diffusion method. The obtained crystals were of sufficient quality for the diffraction experiment on the synchrotron radiation source Bessy II, Helmholtz Zentrum Berlin [3].
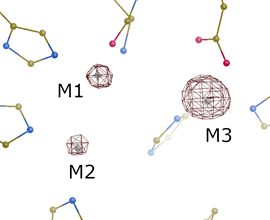
The diffraction data were collected at three different X-ray energies with the aim of detecting the presence of metals using anomalous scattering. Key data collection statistics are summarized in Tab. 1. The obtained anomalous difference maps (Fig. 2) confirmed the exchange of one Zn2+ ion by Ni2+ at the position M3 closest to the enzyme surface, while the other two Zn2+ ions of the core (positions M1 and M2) remained unaffected. Despite the ion exchange, the residues of the active site and its surroundings are structurally conserved.
Table 1: Selected data collection statistics.
|
Space group |
P 212121 |
|
X-ray energy (Ni-low ; Ni-peak ; Zn-peak) [keV] |
8.320 ; 8.346 ; 9.665 |
|
Resolution [Å] |
1.6 |
|
Rmerge (Ni-low ; Ni-peak ; Zn-peak) |
0.072 ; 0.075 ; 0.154 |
|
Mean I/σ (Ni-low ; Ni-peak ; Zn-peak) |
28.8 ; 27.2 ; 15.8 |
|
Avg. anomalous multiplicity |
13 |
|
Completeness [%] |
98.9 |
|
|
|
Figure 1. Secondary structure of S1 nuclease (PDB ID 5FB9). Zinc ions are represented using spheres. Graphics created using PyMOL [4]. |
Figure 2. Anomalous difference map (data set Ni-peak) at a level of 10σ. Significant peak at the position M3 proves the presence of Ni. Graphics created using Coot [5]. |
1. T. Koval’, L. H. Oestergaard, J. Lehmbeck, A. Nørgaard, P. Lipovová, J. Dušková, T. Skálová, M. Trundová, P. Kolenko, K. Fejfarová, J. Stránský, L. Švecová, J. Hašek, J. Dohnálek, PLoS ONE, 11, 2016, e0168832.
2. T. Koval’, J. Dohnálek, Biotechnology Advances, 36, 2018, pp. 603-612.
3. U. Mueller, R. Foerster, M. Hellmig, F. U. Huschmann, A. Kastner, P. Malecki, S. Puehringer, M. Roewer, K. Sparta, M. Steffien, M. Uehlein, P. Wilk, M. S. Weiss. The European Physics Journal Plus, 130, 2015, pp. 141/1-10.
4. L. Schrödinger, W. DeLano, PyMOL, 2020, available from: http://pymol.org/pymol.
5. P. Emsley, B. Lohkamp, W. G. Scott, K. Cowtan, Acta Crystallographica Section D, 66, 2010, pp. 486 – 501.
This work was supported by the MEYS CR (projects CAAS – CZ.02.1.01/0.0/0.0/16_019/0000778 and ELIBIO – CZ.02.1.01/0.0/0.0/15_003/0000447) from the ERDF fund, by the Czech Academy of Sciences (grant No. 86652036), and by the GA CTU in Prague (SGS22/114/OHK4/2T/14). We acknowledge CMS-BIOCEV Crystallization and Diffraction, part of Instruct-ERIC, supported by the MEYS CR (LM2018127).