Breaking the
problem complexity limits for powder diffraction based structure solution
M. Hušák
husakm@vscht.cz
It can be
predicted from theoretical calculations, how much complex structure can be
solved from powder diffraction data [1]. The theoretical limit for perfect
synchrotron data is about 300 DOF (degree of freedom) while current existing record
solves only 42 DOF problem simplified by heavy atom presence [2]. We have tried
to determine a realistic DOF limit based on perfect simulated powder
diffraction data. For the simulation we have chosen peptide structures from CSD:
1 single peptide molecule in asymmetric unit cell, 2-8 amino acids, 10-39 DOF.
The parameters of the simulated powder diffractogram
used were close to typical perfect measurement on ID31 of ESRF - wavelength 0.5
Å, range 0.5º-15º, step 0.002º, FWHM 0.01º. The
structure solution tests were done by SA (simulated annealing) in DASH 3.2
software [3]. To speed up the computation of structures with more than 20 DOF we
have used parallel processing obtained by MDASH extension. Influence of Mogul
CSD based torsion angles bias on the calculation effectiveness was investigated
as well. The results demonstrate the required number of SA steps depends
exponentially on the problems DOF. This requires for problems close to 30 DOF
about 10E+10 SA steps and years of single CPU computational time. The Mogul
based bias can significantly help for compounds like peptides - e.g. for
simulation based on compound CSD code AHAREH (4 peptides, DOF 24) the Mogul
based calculation gives 50 times more often correct result than non-restricted
SA run. We believe the 40 DOF structures can be solved routinely on 16-32 CPU
clusters from perfect data not influenced by preferred orientation when the
Mogul CSD torsion angels biased will be used (required total computational time
about 1 month). Without developing a more efficient algorithm than SA solution
we do not see a way how to get really close to the 300 DOF theoretical limits.
The promising new algorithm can be a brute-force solution space sampling
followed by local minimization as described in [4]. R&D of a code utilizing
this idea is under progress.
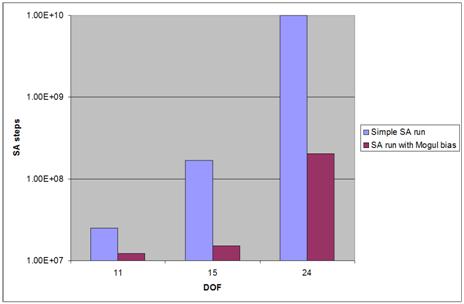
Fig 1.: Dependence of required
simulated annealing steps number required to get one solution on DOF and the
use of Mogul based bias.
1. David,
W.I.F., Shankland, K. (2008). Acta Cryst A64, 52-64.
2. Fernandes, P., Shankland, K., Florence, A. J.,Shankland
N., Johnston A. (2007) Journal of
Pharmaceutical Science 96-5,
1192-1202
3. David, W.I.F., Shankland,
K., Streek J., Pidcock, E.,
Motherwell S., Cole J. (2006). J. Appl. Cryst. 39, 910-915.
4. Shankland K., Markvardsen A.J., Rowlatt
This
work was supported by the Grant Agency of Czech Republic, Grant No.
106/14/03636S.