The structural and biochemical data of enzyme capable of organophosphates degradation
Andrea Štěpánková1,
J. Dušková2, T.
Skálová2,
J. Hašek2, T. Kovaľ3, L. H. Østergaard4, J.
Dohnálek2
1 Dept. of Solid State Physics, FNSPE, CTU, Trojanova 13, 120 00, Prague 2, Czech Republic
2 Institute of Macromolecular Chemistry AS CR, v.v.i., Heyrovského nám. 2, 162 06, Prague 6, Czech Republic
3 Institute of Physic AS CR, Cukrovarnická 10, 162 00, Prague 6, Czech Republic
4 Novozymes A/S, Brudelysvej 26, DK-2880 Bagsvaerd, Denmark
a.stepanko@gmail.com
Extremophiles are organisms living in extreme conditions on Earth (for
illustration: temperatures of
The examined enzyme organophosphorus
acid anhydrolase (OPAA) is able to catalyze hydrolysis of proline dipeptides (Xaa-Pro), and of several
types of organophosphate compounds commonly used as pesticides or as nerve
agents. The enzyme, with the pH optimum around 8, offers a large potential for
biotechnological application as a tool for bio-degradation of these dangerous compounds.
Two different types of the OPAA enzyme from different bacteria – psychrothropic and slightly halophilic Pseudoalteromonas
haloplanktis and slightly halophilic Alteromonas macleodii – were studied and compared with the sequence related
human prolidase.
Three molecular structures of the OPAA
enzyme from Alteromonas macleodii
have been determined. The structure data were
collected at the beam
line PX 14.1 of the source of synchrotron radiation Bessy II (Helmholtz-Zentrum, Berlin).
Native amOPAA crystallized in the space group C2 with unit
cell parameters a =
134.3 Å, b =
49.1 Å, c =
97.2 Å and β =
125.0°. The crystals were measured native and soaked with ligand
Pro-Gly, too. Data were collected up to resolutions 1.8 Å and
1.9 Å, respectively. The third crystal structure determined is the native amOPAA crystallized in the space group P212121 (unit
cell parameters a = 75.6
Å, b = 111.2 Å,
c =
138.1 Å). Data were
collected to resolution 2.2
Å in this case. Refinement of all these
structures performed well with the average B factor around 20 Å2
and the final R factors in the range 15.3 – 16.6 %. The structure of was deposited
in the PDB under the accession code 3RVA.
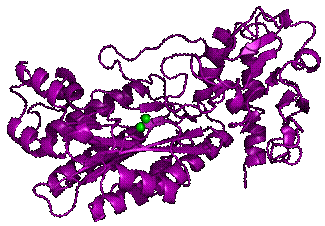
Fig. 1. The cartoon
representation of 3D structure of the
OPAA from Alteromonas macleodii. The
binuclear metal center is highlighted by two spheres.
To
modulate the enzymatic activity profiles and to explain the enzymatic function of
OPAA, we made site-directed mutagenesis of OPAA from Pseudoalteromonas haloplanktis near
in its substrate binding site (Tyr212,
His226, Asp244, Asp255, His335, His339, Arg370, Glu384, Arg421,
Glu423, Val345, His346) found in our structure with the dipeptide
Pro-Gly complex. Only two of seven tested
single-mutations (Y212F, Y212S -
H226N, H226K - H334N, H334K, H334Q) retained the enzymatic activity.
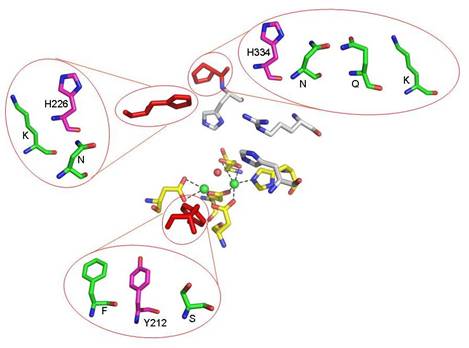
Fig. 2. Tested mutations around the active site
of OPAA. The red residus are in positions found in the native OPAA. The
magenta residues are residues which were mutated in phOPAA; the green residues
show the particular mutations. The manganese ions are shown as green spheres.
Enzyme
assay studies showed a loss of activity as a consequence of site-directed
mutagenesis in five cases. Two mutations highlighted in bold retained the
enzymatic activity according to the results of biochemical characterization
(DLS, SDS-PAGE). The pH profiles of the OPAA mutants Y212F and H226K preserving
activity remain similar to the wild type, but in the case of mutants are shifted
slightly to more basic pH.
The comparative study shows that the OPAA enzyme is very similar to human prolidase. High similarities in structure and also in substrate specificities of OPAAs and prolidases have brought up some interesting questions regarding the historical classification of this group of enzymes with related but not identical activity profiles.
The structures discussed here together with
other three structures of protein-ligand complexes of the low temperature
active β-galactosidase are part of the PhD thesis
/3/.
References:
1.
Vyas, N. K. et al., (2010). Structural insights into the dual activities of the nerve agent degrading organophosphate anhydrolase/prolidase. Biochemistry, 49, 547-559.
2.
Raushel, F. M. (2002). Bacterial detoxification of organophosphate nerve agents. Current opinion in Microbiology, 5, 288-295.
3.
Štěpánková
A., (2012). PhD Thesis, X-ray structural analysis of enzymes from extremophiles
and their complexes with bound ligands, pp. 1-119, FJFI ČVUT, Praha.