STRUCTURE OF
FC-FRAGMENT OF THE MOUSE IMMUNOGLOBULIN
1,3Petr Kolenko,
1Jan Dohnálek, 2Renata Štouračová, 1Tereza
Skálová,
1Galina Tiščenko, 1Jarmila
Dušková, 1Jindřich Hašek.
Mail to: hasek@imc.cas.cz
1 Institute of Macromolecular
Chemistry, Academy of Sciences of CR
2 Institute
of Molecular Genetics, Academy of Sciences of CR
3 Faculty of
Nuclear Sciences and Physical Eng., Czech Technical University of Prague
Immunoglobulines play an irreplaceable role in the immunity system in all higher
organisms. Because of its role in activation of immunological reaction against
infected cells, immunoglobulines are routinely used in
medical applications. There is a very reliable way of IgG
purification based on its affinity to the B fragment of the protein A from Staphylococcus Aureus.
However, the costs of production of protein A are significant and thus a new
method of rapid and cheap production of purified immunoglobulines
in non-denaturating conditions is worth of our special
interest. Any highly selective separation process should be based on molecular
recognition between a specially designed ligand and
the immunoglobulin surface.
In spite of an
immense diversity of Fab fragments containing the hypervariable regions (responsible for molecular
recognition of antibody) at the antigen binding sites, the immunoglobulines
of the same type
(e.g. IgG2b) share very similar aminoacid sequences of
Fc fragment. The object of our interest is Fc-fragment, because of its invariant structure.
Explication of structure of this fragment and its interaction with other
molecules can help in design of polymer sorbents for
affinity chromatography.
Structure determination
Monoclonal antibody,
class IGg2b was cleaved by papain and purified in 4C on
BioLOGIC LP System(BIORAD)
using protein A Sepharose column (Biorad).
The measured crystal was grown under the following conditions:
Reservoir: HEPES pH
7.5, PEG 2000 20 % (w/v). Drop: 1 ml of reseivoir
solution plus 1 ml of protein 8 mg/ml, PBS (phosphate buffer
saline) pH 7.5. Cryoprotectant: 20 % glycerol.
The diffracted
intensities (total 34029 independent reflections) was
collected at the ID-29 beamline at the synchrotron
ESRF in Grenoble with the diffraction limit 2.2
Å. The measured crystal (a triangle platelet 0.3 x 0.1 x 0.02 mm) was
flash cooled to 100 K. Space group is C2, the unit cell a=135,73
Å, b=62,75 Å, c=69,81 Å, β=103,35°. Data reduction was
performed by program package HKL (Denzo, Scalepack, Xdisp)
/1/. The phase problem was solved by molecular replacement (program AMORE /2/)
using the structure model 1IGT taken from PDB /7/ (sequence similarity 79 %). The
other data processing was done mostly using the program package CCP4 /3/. The
structure refinement was done by program REFMAC /4/. Manual corrections were
done by program XtalView /5/, the solvent water
molecules determined with help of ARP/wARP /6/. The methods
used were similar as described in /9/. The final R factors are R=0.19, Rfree=0.25.
The refined structure satisfies all criteria set by program Procheck
/7/ .
Both protein
chains of the Fc fragment IgG2b (residues from Gly125A
to Arg331A and Gly125B to Arg331B) were uniquely identified in the maps of
electron density (except of conformational alternatives evident at side chains
of several residues. The oligosaccharide chains joining the two protein chains
were taken from the protein structure database PDB /8/ - the structure of
immunoglobulin IgG2a (PDB code 1I1C). Both chains -NAG1(FUC)-NAG2-MAN3(MAN-NAG-GAL)-MAN4-NAG5
are chemically bound to Fc fragment through asparagines Asn185A and Asn185B. The
MAN-NAG-GAL branch of both oligosaccharide chains has contacts to its own
protein chain only. The chains -MAN3-MAN4-NAG5
of both Fc fragment halfs
form several hydrogen bonds joining thus both CH2 domains together.
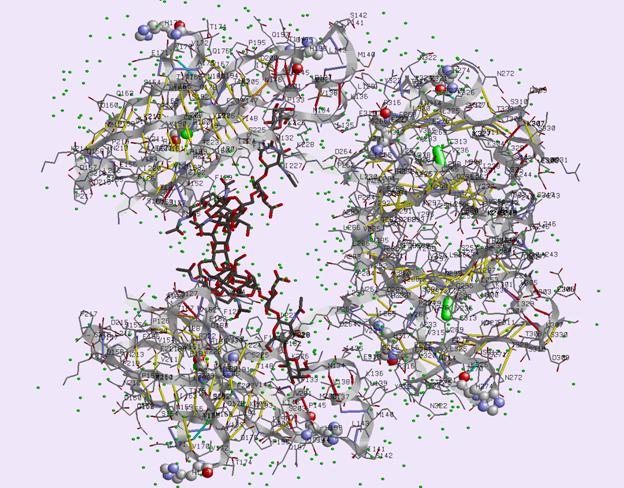
Fig. 1. Schematic view of the Fc fragment of mouse immunoglobulin IgG2b. Each of
the protein chains A,B (upper and lower) divides into
two compact domains Cg2 (loosely joined together by
non-covalent interactions of oligosaccharide chains [black lines] - left side) and
more compact dimer of two Cg3 domains (right side). Small spheres
are water molecules in positions stabilized by hydrogen bridges to the
molecular complex.
Conclusion
A detailed
structure of the intact Fc fragment of IgG2b in
buffer with pH 7.5 determined in this paper is important for elucidation
of the structure changes and the interactions with proteins possessing high
selectivity for Fc fragment surface. These include
complement components responsible for immunological response, viral proteins
(protein A, protein G,...), cell surface receptors and
specially designed molecules suitable for highly efficient separation of immunoglobulines by affinity chromatography.
The project is supported by MSMT - 1K05008.
References
1. Otwinowski Z. & Minor, W. (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzym., 276, 307-326.
2.
Navaza, J. & Saludjian, P. (1994). AMoRe: An
automated molecular replacement program package. Acta Crystallog.Sect. A, 50,
157-163.
3.
Collaborative Computational
Project, Number 4 (1994). The CCP4 Suite: Programs for Protein Crystallography.
Acta Crystallog. Sect. D, 50, 760-763.
4.
Murshudov, G. N., Vagin, A. A. &
Dodson, E. J. (1997). Refinement of Macromolecular Structures by the
Maximum-Likelihood Method. Acta Crystallog. Sect. D, 53, 240-255.
5.
McRee, D. E. (1999). XtalView/Xfit - A Versatile Program for Manipulating
Atomic Coordinates and Electron Density. J.Struct. Biol., 125, 156-165.
6.
Perrakis, A., Harkiolaki, M., Wilson, K.S. & Lamzin, V.S. (2001).
ARP/wARP and molecular replacement. Acta Crystallog. Sect. D, 57, 1445-1450.
7.
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). Procheck - a program to check the
stereochemical quality of protein structures. J. App. Cryst. 26, 283-291.
8.
H.M. Berman, J. Westbrook, Z.
Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne: The
Protein Data Bank. Nucleic Acids Research, 28 pp. 235-242 (2000).
9.
Petroková, H., Vondráčková, E., Skálová, T., Dohnálek, J., Lipovová,
P., Spiwok, V., Strnad,
H., Králová B. & Hašek, J. (2005). Crystallization and preliminary X-ray
diffraction analysis of cold-active b-galactosidase from Arthrobacter sp. C2-2. Collect.
Czech. Chem. C., 70, 124-132.