Database of
Nucleic Acid Structures, NDB
Huanwang
Yang1, Fabrice Jossinet2, Eric Westhof2,
Neocleas Leontis3, Bohdan Schneider4, Chi-Ming Chao1, Zukang
Feng1, Lisa Iype1, Xiang-Jun Lu1, Goran
Aleksic1, Joanna de la Cruz1, Gregory Donahue1,
Dipannita Kalyani1, Daniel Kulp1, Hari Narayan1,
John Westbrook1, and Helen M. Berman1
1Rutgers, The State University of New
Jersey, Department of Chemistry and Chemical Biology, Piscataway, NJ 08854, USA
2Institut de biologie
moléculaire et cellulaire du CNRS, 15 Rue R. Descartes, 676084 Strasbourg,
France
3Chemistry Department, Center for Biomolecular Sciences, Bowling Green State University, Bowling Green OH 43403 USA
4Center for Complex Molecular Systems and
Biomolecules, Dolejskova 3, CZ-18223 Prague, Czech Republic
The Nucleic
Acid Database (NDB) [1, 2] was established in 1991 as a resource of crystal
structures containing nucleic acids. The core of the NDB has been its
relational database of primary and derivative data with very rich query and
reporting capabilities. This robust database was unique in that it allowed
researchers to do comparative analyses of nucleic acid-containing structures
selected from the NDB according to the many attributes stored in the database.
Content of the NDB
Structures available in the NDB include RNA and DNA
oligonucleotides with two or more bases either alone or complexed with proteins
or small molecule ligands. The archive stores both primary and derived
information about the structures. The primary data include: crystallographic or
NMR coordinate data, structure factors for the X-ray structures or contraint
files for the NMR structures, and information about the experiments used to
determine the structures, such as crystallization information, data collection,
and refinement statistics. Derived information, such as valence geometry,
torsion angles, and intermolecular contacts, are calculated and stored in the
database. Database entries are further annotated to include information about
the overall structural features, including conformational classes, special
structural features, biological functions, and crystal-packing classifications.
Data processing and validation
Over the years, the NDB has developed a robust
data-processing system for deposition, processing, archiving, querying, and
distributing structural data. The full capability of this system was recently
demonstrated by the successful processing of ribosomal subunits, which are very
large and complex structures.
This data-processing system is built on top of the mmCIF
dictionary and it naturally supports building of database tables. The NDB has
early on adopted the mmCIF [3] as its data standard. This format has three
advantages from the point of view of building a database: (1) comprehensive definitions
for terms of molecular structure description, crystallography, NMR, as well as electron
microscopy; (2) it is self-defining; and (3) the syntax clearly defines the
relationships between data items. The latter feature is important because it
allows for rigorous checking of internal consistency of the data.
Structures are deposited via the web with the AutoDep Input
Tool (ADIT) [4] and then annotated using the same tool. In the next stage of
data processing, a program called MAXIT (Macromolecular Exchange and Input
Tool) [5] checks and corrects residue and atom naming, numbering, and ordering
as well as the correspondence between the declared sequence and the sequence
based on residue names in the coordinate file. Once these integrity checks are
completed, the structures are validated by NUCheck [6], another program written
as a part of the NDB project. NUCheck verifies valence geometry, torsion
angles, intermolecular contacts, and the chiral centers of the (deoxy)riboses
and phosphates.
The NDB query capabilities
The core of the NDB project is a relational database in
which all data items are organized into tables. At present, there are over 90
tables in the NDB, with each table containing 5 to 20 data items. These tables
contain both experimental and derived information. Example tables include: the
citation table, the cell dimension table, and the refine parameters table.
Interaction with the database is a two-step process. In the
first step, the user defines the selection criteria by combining different
database items. Once the structures that meet the constraint criteria have been
selected, reports may be written using a combination of table items. For any
set of chosen structures, a large variety of reports may be created, e.g. a
crystal data report, a backbone torsion angle report, or the user could write a
report that lists the twist values for all CG steps in the selected structures together
with statistics, including mean, median, and range of values. An important
feature of the NDB capabilities is that the constraints used for the reports do
not have to be the same as those used to select the structures.
The changing face of the ndb
Since the NDB project began in early 1990s, our knowledge of
nucleic acid structures has grown in quantity as well as quality. Early structures
of DNA and RNA oligonucleotides, a few protein-DNA complexes, and some tRNA
structures available in 1990, have been extended by hundreds of protein – DNA
complexes, ribozyme structures and the newest additions to the archive—ribosomal
subunit structures. Not of the least importance is the growth of nucleic acid
structures solved by NMR techniques.
All this had to be reflected by
changes in the NDB itself. During the last three years, the NDB is undergoing a
gradual change. The changes started by unifying data structure and adding new
data items and corresponding database tables. Further, over 500 NMR structures
have been added to the NDB. These changes offered to build new query tools and
possibilities for more flexible and reliable searches. A new web interface was
designed to make the query capabilities of the NDB as widely accessible as
possible and easy to use. Figure 1 shows the new NDB home page, with
possibility for ID and keyword search and key links.
Implementation of these changes
required a complete overhaul of the layout of the NDB web pages including new
style for all graphic representation of nucleic acids (Lu & Olson, in
preparation; [7]). Figure 2 illustrates some
possibilities of the new searches.

Figure 1. NDB home page, http://ndbserver.rutgers.edu.
The largest changes have been
made to the Atlas pages which have been completely overhauled. The
classification of structures have been simplified, structures are presented in
thumbnail galleries, an example for ribozyme structures is shown in Figure 3,
and design of the pages for individual
structures have also been modified to give the user a more informative overview
of a structure (Figure 4).
In the new Search mode, several items, including structure
ID, author, and several classification and structural features can be limited
either by entering text in a box or by selecting a Yes or No option. Any
combination of these items may be used to constrain the structure selection. If
none are used, the entire database will be selected. After selecting “Execute
Selection” the user will be presented with a list of structure IDs and
descriptors that match the desired conditions. Several viewing options for each
structure in this list are possible. These include retrieving the coordinate
files in either mmCIF or PDB format, retrieving the coordinates for the
biological unit, or viewing an NDB Atlas page. Preformatted Quick Reports can
then be generated for the structures in this result list. Multiple reports can
be easily generated, e.g. to get bibliographic information, refinement
statistics, and backbone torsion angles for the selected structures.
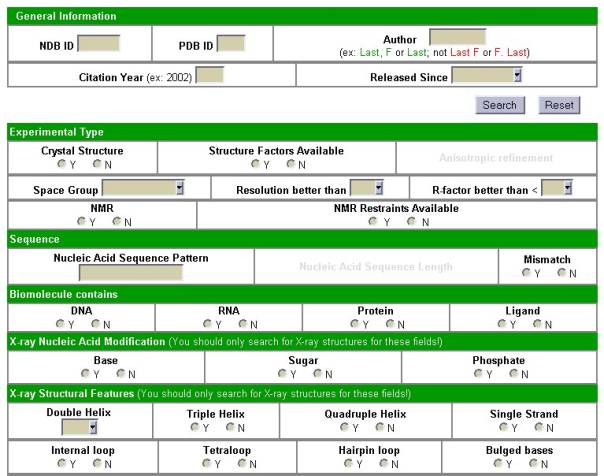
Figure 2. Some search options of
the NDB.
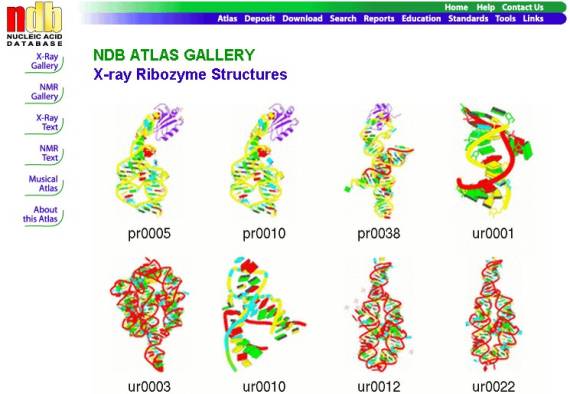
Figure 3. NDB Atlas gallery for ribozyme crystal structures.
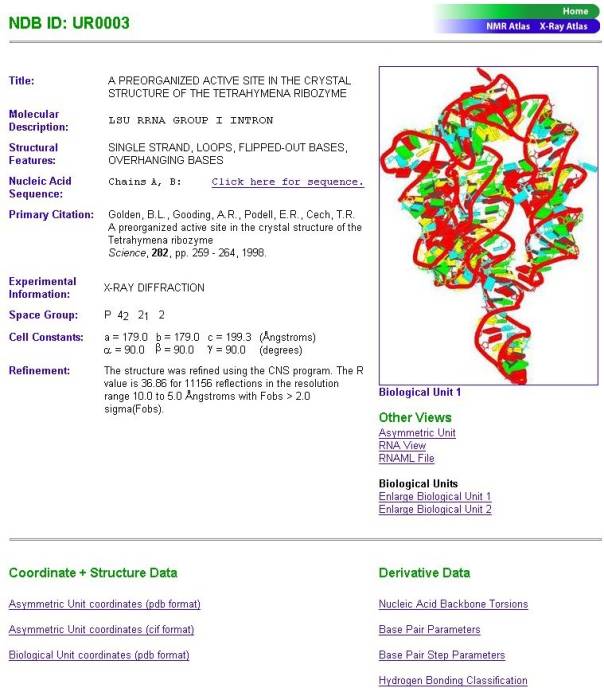
Figure 4. Atlas page of the structure , NDB code UR0003.
In the Full Search/Full Report mode, it is possible to
access most of the tables in the NDB to build more complex queries. Instead of
limiting items that are listed on a single page, the user builds a search by
selecting the tables and then the items that contain the desired features.
These queries have selectable Boolean and logical operators to make complex
queries. After selecting structures using the Full Search, a variety of reports
can be written. The report columns are selected from a variety of database
tables, similar to the tables used for the Full Search.
Data distribution
Coordinate files, database reports, software programs, and
other resources are available via the ftp server (ftp://ndbserver.rutgers.edu).
In addition to links to information provided from the ftp server, the web
server (http://ndbserver.rutgers.edu/) provides a variety of methods for
querying the NDB. These sites are updated continually.
Acknowledgements
The NDB Project is funded by the National Science Foundation
and the Department of Energy. BS is supported by a grant from the Ministry of
Education of the Czech Republic No. LN00A032 for the Center for Complex
Molecular Systems and Biomolecules.
References
[1] Berman H.M., Olson W.K., Beveridge D.L., Westbrook J.,
Gelbin A., Demeny T., Hsieh S.-H., Srinivasan A.R., Schneider B. (1992): The
Nucleic Acid Database—a comprehensive relational database of three-dimensional
structures of nucleic acids. Biophys. J. 63:751–9. [This paper gives the full
description of the NDB system.]
[2] Berman H.M., Feng Z., Schneider B.,Westbrook J.,
Zardecki C. (2001): The Nucleic Acid Database (NDB). In: Rossman M.G., Arnold E.,
editors. International Tables for Crystallography, F. Crystallography of
Biological Macromolecules. Dordrecht: Kluwer Academic Publishers. pp. 657–662.
[3] Bourne P.E., Berman H.M., Watenpaugh K., Westbrook J.D.,
Fitzgerald P.M.D. (1997): The macromolecular Crystallographic Information File
(mmCIF). Methods Enzymol. 277:571–90.
[4] Westbrook J., Feng Z., Berman H.M. (1998): ADIT—The
AutoDep Input Tool. Department of Chemistry, Rutgers, the State University of
New Jersey, RCSB-99.
[5] Feng Z., Hsieh S.-H., Gelbin A., Westbrook J. (1998a):
MAXIT: Macromolecular Exchange and Input Tool. New Brunswick, NJ: Rutgers
University, NDB–120.
[6] Feng Z., Westbrook J., Berman H.M. (1998b): NUCheck. New
Brunswick, NJ: Rutgers University, NDB–407.
[7] Yang, H., Jossinet,
F., Leontis, N., Chen, L., Westbrook, J., Berman, H. M., and Westhof, E. (in
press).