X-RAY STRUCTURES
OF SELECTED N6-SUBSTITUTED ADENINE DERIVATIVES AND THEIR COPPER(II)
COMPLEXES
M. Maloň1,
M. Šipl1, Z. Trávníček*1 and J. Marek2
1Department of Inorganic Chemistry, Faculty of Science, Palacký
University, CZ-771 47 Olomouc, Czech Republic, e-mail*: trav@aix.upol.cz
2X-ray Laboratory of Faculty of Science, Masaryk University, CZ-611
37 Brno, Czech Republic
In general, N6-substituted adenine derivatives belong to a class of natural and artificial plant growth regulators called cytokinins. Their dominant physiological effect is to induce cell division [1], even if the molecular mechanisms by which cytokinins mediate cell growth and division are still unknown. Recently, it has been discovered that some of cytokinin-derived compounds, e.g. Olomoucine and related compounds, specifically inhibit some cyclin-dependent kinases at micromolar concentration [2]. Moreover, it has been found that these compounds also show cytotoxic activity against some of human cancer cell lines. This biological activity can be improved by interaction of these organic compounds with selected transition metals [3,4].
As part of our systematic study of purines bearing C-substituents in positions N6, C2 and/or N9 and their transition metal complexes, we have prepared and structurally characterized the following compounds: 6-(3-chlorobenzylamino)purinium chloride (1), 6-(4-methoxy-benzylamino)purinium chloride (2), [Cu(2MeObapH+-N9)2(H2O)Cl2]Cl2.H2O (3) and [Cu(2FbapH+-N9)2Cl3]Cl.(H2O)2 (4), where 2MeObap = 6-(2-methoxybenzylamino)purine and 2Fbap = 6-(2-flourobenzylamino)purine. Molecular structures of the compounds 1-4 are depicted in Figures 1-4. The cytotoxic activity study of the Cu(II)-complexes against G-361 (human malignant melanoma), HOS (human osteogenic sarcoma), K-562 (human chronic myelogenous leukemia) and MCF-7 (human breast adenocarcinoma) cell lines is the subject of our present investigation. Here, we describe only preliminary crystallographic results.
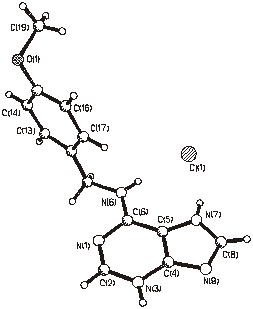
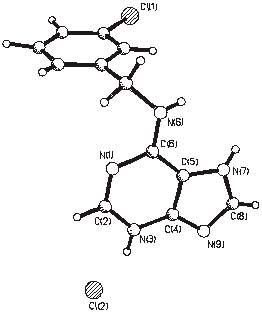
Figure 1. Molecular structure of (1). Figure 2. Molecular structure of (2).
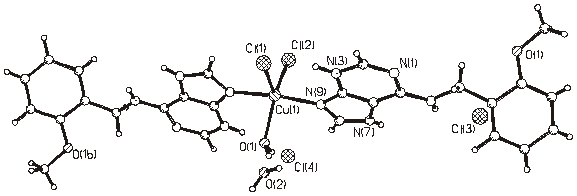
Figure 3. Molecular structure of (3).
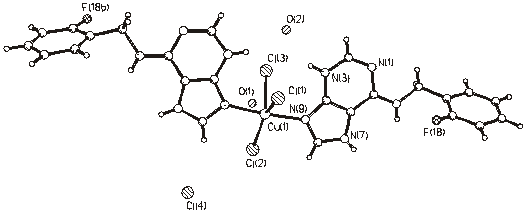
Figure 4. Molecular structure of (4).
X-ray data for all compounds were collected at 120(2) K on a four-circle kappa-axis diffractometer KUMA KM-4 equipped with an Oxford Cryostream cooler and a CCD detector. All crystal structures were determined and refined using a SHELX97 program package.
Crystal data and refinement for (1): C12H11N5Cl2, monoclinic P21/c, a = 18.6316(12), b = 8.8554(5), c = 8.0320(5) Ĺ, b = 96.219(6)°, Z = 4, F(000) = 608, GOF = 1.004,
reflections collected / unique 6760 / 2282 [Rint = 0.0341], R1obs = 0.0640, wR2obs = 0.1419.
Crystal data and refinement for (2): C13H14N5Cl1O1, triclinic P-1, a = 4.9572(11), b =
9.260(2), c = 14.453(3) Ĺ, a = 90.95(2), b = 94.89(2), g = 100.34(2)°, Z = 2, F(000) = 304,
GOF = 0.855, reflections collected / unique 4070 / 2224 [R(int) =
0.0554], R1obs = 0.0378, wR2obs = 0.0980.
Crystal data for and refinement (3): C26H31N10Cl4CuO4, monoclinic Cc, a = 10.8038(4), b = 10.1711(4), c = 28.3908(10) Ĺ, b = 90.596(3)°, Z = 4, F(000) = 1544, GOF = 1.029,
reflections collected / unique 6139 / 3568 [Rint = 0.0397], R1obs = 0.0343, wR2obs = 0.0879.
Crystal data for and refinement (4): C24H24N10Cl4CuF2O2, monoclinic P21/n, a = 14.8791(5), b = 8.3991(3), c = 23.5003(8) Ĺ, b = 95.109(3)°, Z = 4, F(000) = 1476, GOF = 1.034,
reflections collected / unique 15265 / 5120 [Rint = 0.0652], R1obs = 0.0418, wR2obs = 0.1092.
Acknowledgement: This work was financially supported by the Grant Agency of the Czech Republic (203/00/0152).
References
- D.S. Letham & L.M.S. Palni, Annu. Rev. Plant. Physiol., 34 (1983) 163-167.
- J. Veselý, L. Havlíček, M. Strnad, J.J. Blow A.
Donella-Deana, L. Pinna, D.S. Letham, J.Y. Kato, L. Detivaud, S. Leclerc & L. Meijer, Eur. J. Biochem., 224
(1194) 771-786.
- Z. Trávníček, M. Maloň, Z. Šindelář, K. Doležal, J. Rolčík, V. Kryštof, M. Strnad & J. Marek, J. Inorg. Biochem., 84 (2001) 23-32.
- M. Maloň, Z. Trávníček, M. Maryško, R. Zbořil, M. Mašlán, J. Marek, K. Doležal, J. Rolčík, V. Kryštof & M. Strnad, Inorg. Chim. Acta, 323 (2001) 119-129.