Biophysical and binding characterization of rhesus CMV UL144 as a promising modulator of the T-lymphocyte CD160 pathways
A. Bitala1, M. Benko1, S. Lenhartová1, M. Mladá1, M. Nemčovič2, I. Nemčovičová1
1Biomedical Research Center, Institute of Virology, Slovak Academy of Sciences, Bratislava, Slovakia
2Institute of Chemistry, Slovak Academy of Sciences, Bratislava, Slovakia
andrej.bitala@savba.sk
Viral immunomodulatory glycoprotein UL144 isolated from Rhesus Cytomegalovirus (RhCMV) is homologous to human Cytomegalovirus (HCMV) UL144 and highly orthologous to human tumour necrosis factor receptor HVEM (TNFR/SF14, herpesvirus entry mediator). Endogenous HVEM
functions in bi-molecular switch to regulate the host's immune response depending on which ligand is currently binding [1]. Despite the high structural similarities between UL144 viral glycoproteins and the HVEM, they do not share the same binding properties therefore could modulate the immune response
differently. An example is the activation of the inhibitory pathway of T-lymphocytes through the coinhibitory molecule CD160 (the natural killer cell–activating receptor). This molecular network is quite well described, but the engagement of CD160 by UL144 has not been satisfactorily studied yet [2]. We
are currently focusing on RhCMV UL144, which predicted to interact with both human and the rhesus CD160 with a low affinity [3]. Such evidence could represent the evolutionary divergence between viral species.
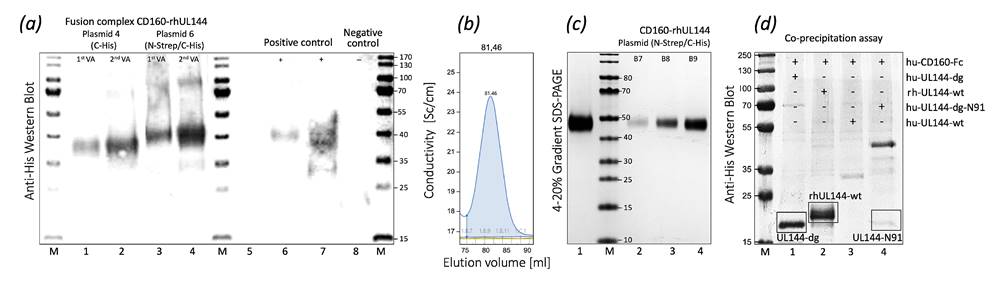
Figure 1. Expression, purification, and binding characteristics of huCD160-rhCMVUL144 fusion complex monitored by Western blot (a), FPLC (b), SDS-PAGE (c) and co-precipitation assay (d).
To fully understand the molecular basis of the interactions between human CD160 and RhCMV UL144,we created their covalently linked protein complex via a baculovirus-mediated expression systém in Spodoptera frugipherda (BV-Sf9) insect cells. The plasmid pAcGP67A was used as a transfer vector,
into which the synthetically prepared gene for the fusion complex was cloned in the multi-cloning side. Other variants of CD160 and UL144 alone (e.g., UL144-WT, UL144-DG, UL144-N91, CD160-Fc) were similarly prepared and expressed in BV-Sf9. By performing the standard WB with HRPconjugated antibody interacting with the protein’s affinity tags we clearly identified the expressed proteins. The protein complex huCD160–rhCMV UL144 was further analyzed by gel filtration chromatography and SDS-PAGE methods and the molecular size of the complex was confirmed (Fig. 1). Furthermore, the molecular mass for UL144 variants were determined by MS. To determine the stabilization characteristics of the purified proteins, the melting temperature (Tm) was estimated biophysically by using the Nanotemper device. The obtained Tm was lower in glycosylation mutants,
and onset of unfolding temperature. We also specified the quality of these samples with further measurements of polydispersity index, hydrodynamic radius, and turbidity. In addition, we attempted to confirm the binding capacity of RhCMV UL144 by the co-immunoprecipitation assay where the binding
to CD160 was observed. Moreover, all formed complexes and individual proteins were attempted to crystallize by vapor-diffusion technique in a sitting-drop using commercially available crystallization screens. All obtained data will be further analyzed and optimized.
1. C. F. Ware CF, and J. R. Šedý, Curr. Opin. Immunol. 23, (2011), 627-631.
2. A. Bitra, I. Nemčovičová, et al. and D. Zajonc, J. Biol. Chem. 294, (2019), 10519–10529.
3. J.R. Šedý, R.L. Bjordahl, et al. and C.F. Ware, J. Immunol. 191, (2013), 828–836.
Financial supports provided by the Slovak Research and Development Agency (APVV-19-0376) and
the Scientific Grant Agency of the Slovak Republic (VEGA-02/0026/22 and VEGA-02/0060/21) are gratefully acknowledged.