Mutational analysis on viral HCMV UL141 protein to specify the binding site of novel antagonist that blocks TRAIL-R2 binding
I. Nemčovičová1, J. Kóňa2, M. Poláková2, T. Klunda2, A. Bitala1, M. Benko1, and M. Nemčovič2
1Biomedical Research Center, Slovak Academy of Sciences, Bratislava, Slovakia
2Institute of Chemistry, Slovak Academy of Sciences, Bratislava, Slovakia
viruivka@savba.sk
The human cytomegalovirus (HCMV) worldwide seroprevalence is estimated to 83% in the general population [1-2]. Usually, it is controlled by a vigorous immune response so that infections are asymptomatic, or symptoms are mild. However, if the immune system is compromised, HCMV can replicate to high levels and cause serious end organ disease [3]. Although HCMV represents a complex target, the rounds of iterative studies could potentially bring this important and under-recognized human pathogen under control.
Over four receptor-binding patches (RBP) of the glycosylated viral protein UL141 is capable of binding to human TRAIL death receptor 2 (TRAIL-R2) [4] and trigger the NK signalling pathway to benefit viral fitness [5-7]. Hence, it is rational to inhibit the RBP activity of the UL141 protein by blocking the RBP interaction with TRAIL-R2, which makes the UL141 a potential target for designing and developing antiviral agents. In this study, the molecular features of the UL141 of HCMV are highlighted, such as the structure, functions, and interactions of the UL141 and TRAIL-R2. Furthermore, the development of glycomimetic structures by computational design and biochemical testing is reported.
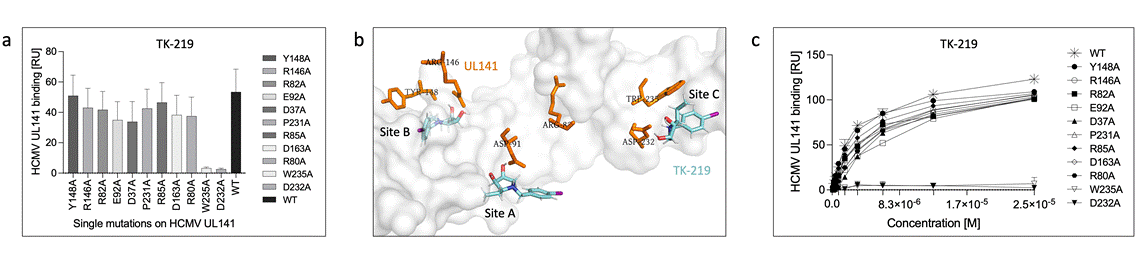
Figure 1. Three potential binding sites were identified by molecular docking on UL141 surface (b). Mutational analysis revealed the specific binding site of novel antagonist TK-219 (a). SPR binding analysis (c) showed two UL141 mutants that lost the ability to block the TRAIL-R2 binding.
The aim is to develop the short peptide or synthetic compound (UL141 antagonist) based on our recent crystal structure and computational design that would specifically bind viral UL141 to block receptor binding thus prevent the viral action. This is relevant, as the UL141 is also the most abundant HCMV protein on plasma membrane and it is also a component of the virion. Based on our computational screening of iminosugars the 'hit' structure was selected. We test a small library of synthetized compounds (potential UL141 antagonists) that would block the receptor binding in vitro, on the cell or virion surface. Series of compounds that have been tested are of glycomimetics structures consisting of various saccharide units linked with non-saccharide.
In particular, non-ionic glycolipids, 'click'-conjugates or iminosugars. The ELISA-like TMB assay has been used in combination with dynabeads coating to test whether the compound could block the TRAIL-R2 binding. Five most promising compounds have proven the ability to block UL141/TRAIL-R2 complex formation. SPR kinetics analysis was then used to determine the binding constants (KD). The affinities to UL141 were determined in low micromolar scale. Three potential binding sites were revealed by molecular docking on UL141 surface (Fig. 1b). Next, mutational analysis on UL141 protein (Fig. 1a) has revealed the specify binding site of novel antagonist TK-219. Subsequent SPR binding analysis (Fig. 1c) showed two UL141 mutants (W235A and D232A) lost the ability to block the TRAIL-R2 binding. The successful compounds will be further optimized by using in silico methods to target epitope on viral glycoprotein UL141 derived from our structural analysis and will be tested in vivo for HCMV inhibition.
1. Zuhair, M. et al. Rev. Med. Virol., 29, (2019), e2034.
2. Picarda, G. & Benedict, C. A. J. Immunol., 200, (2018), 3881.
3. Nemčovičová, I., Benedict, C. A. & Zajonc, D. M. PLoS Pathog., 9, (2013), 1003224.
4. Nemčovičová, I. & Zajonc, D. M. Acta Crystallogr. D Biol. Crystallogr., 70, (2014), 851.
5. Smith, W., Tomasec, P., Aicheler, R., Loewendorf, A. Nemčovičová, I. et al. Cell Host Microbe., 13, (2013), 324.
6. Tomašec, P., Wang, E. C. Y., Davison, A. J., Vojtešek, B., Armstrong, M. et al. Nat. Immunol., 6, (2005), 181.
Financial supports provided by the Slovak Research and Development Agency (APVV-19-0376) and the Scientific Grant Agency of the Slovak Republic (VEGA-02/0026/22 and VEGA-02/0060/21) are gratefully acknowledged.