New Scramblases with Moonlighting Functions Identified in Mitochondria
L. Barto1,2, H. Jahn3, A.K. Menon3, R. Vácha*1,2,4
1CEITEC -- Central European Institute of Technology, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic
2National Centre for Biomolecular Research, Faculty of Science, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic
3Department of Biochemistry, Weill Cornell Medical College; New York, NY 10065, USA
4Department of Condensed Matter Physics, Faculty of Science, Masaryk University, Kotláĝská 2, 611 37 Brno, Czech Republic
robert.vacha@muni.cz
Mitochondria are the essential powerhouses of eukaryotic cells, and their optimal function depends on a specific phospholipid composition in their two membranes. While some of these lipids are supplied by the endoplasmic reticulum, the mechanism that facilitates their transfer across the outer mitochondrial membrane has been elusive. Our research has identified novel scrambling proteins responsible for lipid transport across the outer mitochondrial membrane.
The first identified protein is the voltage-dependent anion channel (VDAC), an abundant component of the mitochondrial outer membrane [1]. Although VDAC is widely recognized for its ion channel activity, our research has revealed its previously unknown role as a phospholipid scramblase. This novel function was demonstrated with in vitro experiments using yeast mitochondria and reconstituted vesicles, and is further supported by coarse-grained molecular dynamics simulations of VDAC [2]. These simulations have elucidated the mechanism of lipid scrambling, which occurs at specific interface between VDAC dimers (Figure 1a). This interface contains polar residues that create significant water defects and thin the lipid bilayer, facilitating lipid scrambling. VDAC, with its beta-barrel transmembrane structure, introduces a new class of phospholipid scramblases [2], which are distinct from previously identified scramblases with alpha-helical transmembrane structures.
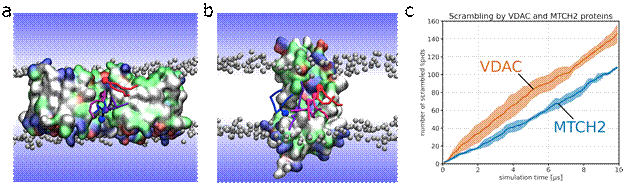
Figure 1. Lipid scrambling by VDAC and MTCH2 proteins. Snapshots of VDAC dimer a) and MTCH2 b) from coarse grained simulations depict phospholipids translocating between membrane leaflets along the scrambling pathway at the protein. The translocating lipids are colored in red, purple, and blue. Proteins are shown as a molecular surface, with colors indicating the character of its residues (hydrophilic = green, hydrophobic = white, positively charged = blue, negatively charged = red). Lipids of bulk membrane are depicted as gray beads (phosphate groups) with hydrophobic tails omitted for clarity. Water is represented only schematically as a blue gradient. c) Scrambling rate of VDAC dimer and MTCH2 proteins in a POPC membrane are shown as the number of scrambled lipids in time. Lipid was considered scrambled when it was present in the opposite leaflet than its original one.
The second identified protein, MTCH2, is also located in the mitochondrial outer membrane and has previously been recognized as an insertase [3]. Unlike VDAC, MTCH2 is characterized as a helical transmembrane protein similar to previously identified scramblases. MTCH2 has a hydrophilic groove in the transmembrane region that locally thins the hydrophobic core of the membrane and facilitates the lipid scrambling (Figure 1b), similarly to VDAC [4]. Molecular dynamics simulations, both coarse-grained and atomistic, were used to demonstrate that the groove is responsible for lowering the free energy barrier for lipid movement across the membrane. The scrambling rate observed for MTCH2 is comparable to that of VDAC (see Figure 1c), suggesting its potential complementary role in mitochondrial lipid transport. Although VDAC and MTCH2 have distinct structural features, they share a common mechanism of facilitating lipid transport by locally thinning the membrane [4].
These discoveries represent a significant advancement in our understanding of lipid transport in mitochondria. In addition, the dual functionality of these proteins highlights the complexity of mitochondrial biology and provides opportunities for further research in membrane dynamics and cellular metabolism.
1. Bergdoll, L.; Grabe, M.; Abramson, J.: An Assessment of How VDAC Structures Have Impacted Our Understanding of Their Function. Molecular Basis for Mitochondrial Signaling, 2017, Springer International Publishing AG, 141-160.
2. Jahn, H.; Barto, L.; Dearden, G.I.; Dittman, J.S.; Holthuis, J.C.M.; Vácha, R.; Menon, A.K.: Phospholipids are imported into mitochondria by VDAC, a dimeric beta barrel scramblase. Nature Communications 2023, 14, 8115.
3. Guna, A.; Stevens, T.A.; Inglis, A.J.; Replogle, J.M.; Esantsi, T.K.; Muthukumar, G.; Shaffer, K.C.L.; Wang, M.L.; Pogson, A.N.; Jones, J.J.; Lomenick, B.; Chou, T.F.; Weissman, J.S.; Voorhees, R.M.: MTCH2 is a mitochondrial outer membrane protein insertase. Science 2022, 21, 378(6617), 317-322.
4. Barto, L.; Menon, A.K.; Vácha, R.: Insertases Scramble Lipids: Molecular Simulations of MTCH2 2024, Structure, in press.
Acknowledgements: This work was supported by the National Institutes of Health grant NS119779 (AKM), the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement No 101001470) (RV) and the project National Institute of virology and bacteriology (Programme EXCELES, ID Project No. LX22NPO5103) - Funded by the European Union - Next Generation EU (RV).
Computational resources were provided by the CESNET, CERIT Scientific Cloud, and IT4 Innovations National Supercomputing Center by MEYS CR through the e-INFRA CZ (ID:90254). We acknowledge IT4 Innovations National Supercomputing Center for awarding this project access to the LUMI supercomputer, owned by the EuroHPC Joint Undertaking, hosted by CSC (Finland) and the LUMI consortium through the e-INFRA CZ (ID:90254).