Exchanging Metals in Zn-Dependent S1 Nuclease: Effects on Structure and Activity
J. Hrubý1,2, P. Kolenko1,2, K. Adámková2,3, B. Husťáková2,3, M. Malý1,2,4, L. H. Østergaard5, T. Kovaľ2, J. Dohnálek2
1Czech Technical University in Prague, Břehová 7, 115 19 Prague, Czech Republic
2Institute of Biotechnology of the Czech Academy of Sciences, Biocev, Průmyslová 595, Vestec,
3University of Chemical and Technology Prague, Technická 5, Prague, Czech Republic
4Institute for Life Sciences, University of Southampton, Southampton, SO17 1BJ, United Kingdom
5Dept. of Agile Protein Screening, Novozymes A/S, Krogshoejvej 36, Bagsvaerd, Denmark
Corresponding authors: hrubyj18@fjfi.cvut.cz (JH); jan.dohnalek@ibt.cas.cz (JD)
Metalloenzymes constitute a large portion of known enzymes. The presence of metals in the active site of those enzymes can be crucial for both enzymatic activity and structural integrity [1].
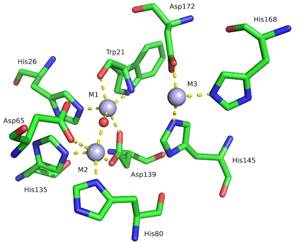
S1 nuclease from Aspergillus oryzae is a Zn-dependent, single-strand specific enzyme with wide utilization in the biotechnology industry and research [2,3]. The active site of S1 consists of a zinc cluster containing three Zn2+ ions and 9 surrounding amino acid residues coordinating the cluster (Fig. 1). The shape of the active site resembles a pocket: Two Zn2+ ions (M1, M2) are buried at the bottom while the third Zn2+ ion is located at the pocket opening (M3), closer to the surface of the nuclease.
We studied the consequences of replacing Zn with various metals on the structure of the active site and the enzymatic activity towards the ssDNA substrate. For our work, S1 was first treated with a chelating agent ethylenediaminetetraacetic acid (EDTA). After the addition of various metals (CuCl2, CdCl2, FeCl3, and NiCl2), the resulting mixture was crystallised using the vapour diffusion method. Subsequently, X-ray diffraction experiments using multiple X-ray energies were conducted at Bessy II, Helmholtz Zentrum Berlin [4]. The activity towards ssDNA as a substrate was measured using precipitation of undigested nucleic acids and measurement of absorbance at 260 nm.
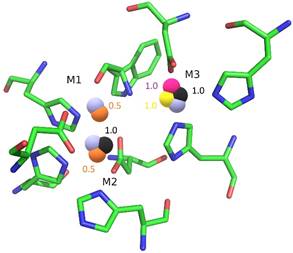
Using anomalous scattering at different X-ray energies, we proved that it is possible to artificially replace the Zn atoms in the active site (Fig. 2). Moreover, we discovered that all three positions are prone to exchange, with position M3 being the most frequent (Cu, Cd, and Ni were observed at this position). In all four cases, the exchange was incomplete with a maximum of two atoms replaced at a time, the remaining positions were occupied by Zn. The structure of the active site remained well conserved in comparison with native S1 (PDB ID 5FB9, [2]) with no significant structural changes.
The activity studies showed that, despite little to no structural changes, S1 remained inactive after the EDTA treatment and subsequent metal ion replacement using Cu, Cd, Fe and Ni. This suggests that the presence of three Zn ions is crucial for the activity and the metal replacement itself is sufficient for the complete inhibition of enzymatic activity.
1. C. Dupureur, Current Opinion in Chemical Biology, 12(2), 2008, pp. 250-255.
2. T. Koval’, L. H. Oestergaard, J. Lehmbeck, A. Nørgaard, P. Lipovová, J. Dušková, T. Skálová, M. Trundová, P. Kolenko, K. Fejfarová, J. Stránský, L. Švecová, J. Hašek, J. Dohnálek, PLoS ONE, 11, 2016, e0168832.
3. T. Koval’, J. Dohnálek, Biotechnology Advances, 36, 2018, pp. 603-612.
4. U. Mueller, R. Foerster, M. Hellmig, F. U. Huschmann, A. Kastner, P. Malecki, S. Puehringer, M. Roewer, K. Sparta, M. Steffien, M. Uehlein, P. Wilk, M. S. Weiss. The European Physics Journal Plus, 130, 2015, pp. 141/1-10.
5. L. Schrödinger, W. DeLano, PyMOL, 2020, available from: http://pymol.org/pymol.
Figure 1. Active site of native S1 nuclease (PDB ID 5FB9, [2]). Zinc atoms and water molecule are represented using spheres in light blue and red, respectively. The surrounding structure is represented using sticks (C green, O red, N blue). Molecular graphics were created using PyMOL [5]. |
Figure 2. Schematic representation of all metal ion replacements in the active site. The position M3 (top right) is the most frequently replaced. Metal atoms are represented using spheres: Cu in pink, Cd in black, Fe in orange, Ni in yellow, and Zn (natively present in the active site) in light blue. Numbers represent the occupancy of substituent metals. Labels of the residues are omitted for clarity. Molecular graphics were created using PyMOL [5]. |
This work was supported by the MEYS CR (projects CAAS – CZ.02.1.01/0.0/0.0/16_019/0000778 and ELIBIO – CZ.02.1.01/0.0/0.0/15_003/0000447) from the ERDF fund, by the Czech Academy of Sciences (grant No. 86652036), and by the Grant Agency of the Czech Technical University in Prague, grant No. SGS22/183/OHK4/3T/14. We acknowledge CMS-BIOCEV Crystallization and Diffraction, part of Instruct-ERIC, supported by the MEYS CR (LM2018127).