Structural study of RNA-preferring nuclease SmNuc1 from Stenotrophomonas maltophilia
K. Adámková1, 2, T. Kovaž1, M. Trundová1, B. Husáková1, J. Dohnálek1
1Institute of Biotechnology of the Czech Academy of Sciences, v.v.i., Průmyslová 595, 252 50 Vestec, Czech Republic
2 University of Chemistry and Technology Prague, Department of Biochemistry and Microbiology, Technická 5, 166 28 Prague 6, Czech Republic
adamkovak@ibt.cas.cz
Zinc-dependent nuclease from pathogenic bacterium Stenotrophomonas maltophilia (SmNuc1) is a highly active RNA-preferring nuclease from the S1/P1 family (EC 3.1.30.1). Members of this family, especially from fungi and plants, have already been characterized and their structures were solved [1], but many questions remained unanswered. As they are widely used in biotechnology and biochemistry, new, more detailed information on their cleavage mechanism, substrate preferences, and active site composition may lead to an expansion of their potential applications.
Here we present a structure of recombinant SmNuc1 at 1.4 Å resolution followed by high-resolution (1.20 - 1.85 Å) structures of complexes with RNA cleavage products (mononucleotides). These complexes show possible binding of products to the active site of SmNuc1 after cleavage and also their binding in novel, inhibitor-like modes. Also, we were able to capture some intermediate states of RNA cleavage process (product leaving), and this helped us to suggest a binding mode of longer RNA oligonucleotide substrates. Interestingly, close to the active site we detected a mobile segment, previously unseen in this family, containing the active‑site‑forming Arg74 and capable of remodeling the substrate binding site, which raises some new questions, such as is this remodeling the main mechanism for substrate specificity?
Structural and kinetic studies of SmNuc1 brought new insights into the cleavage mechanism and in combination with the already known facts about S1/P1 nucleases we are closer to clarifying some aspects of substrate preferences.
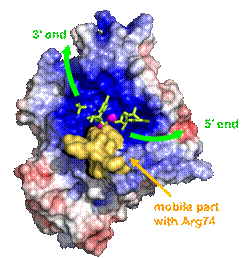
Figure 1: SmNuc1 surface coloured by electrostatic potential from red (−) to blue (+). The yellow surface indicates the mobile segment with Arg74 close to the substrate binding site. Green arrows indicate the orientation of nucleic acid binding. Ligands are shown in green sticks and zinc ions as violet spheres. Graphics was created using PyMOL (Schrödinger).
1. Kovaž T, Dohnálek J, Biotechnology Advances, 2018, 36(3): 603-612
The work was supported by the institutional support of IBT CAS, v.v.i. (RVO: 86652036), CSF (23-06295S), ERDF (CZ.02.1.01/0.0/0.0/15_003/0000447, CZ.02.1.01/0.0/0.0/16_013/0001776 and CZ.1.05/1.1.00/02.0109), MEYS CR (CZ.02.1.01/0.0/0.0/16_019/0000778 and LM2018127, support of Biocev-CMS core facilities Crystallization of Proteins and Nucleic Acids, and Structural Mass Spectrometry of CIISB, part of Instruct-ERIC).