What governs protein:protein interactions of C-type lectin-like domains?
J. Dohnálek, T. Skálová
Institute of Biotechnology, Czech Academy of Sciences, Průmyslová 595, 25250 Vestec
dohnalek@ibt.cas.cz
Protein domains with the C-type lectin-like (CTL) fold are widely utilized in important protein:protein interactions. The molecular and cellular context of these interactions covers various systems from snake venoms to human immune system components. The protein:protein interaction affinity for the individual cases differs within six orders of magnitude (!) and all the possible types of protein:protein interactions (hydrogen bonds, shape complementarity etc.) are being used.
For analysis of the CTL-based protein:protein interactions, we used a specific method: mapping of interactions onto one CTL representative (human CD69). This enabled comparison of individual cases of CTL surface utilization and definition of typical classes [1]. Several types of interactions of CTL domains can be distinguished, none of them requiring strict surface sequence conservation. The CTL interaction classification brings a better understanding of this frequently used domain type and draws the basic framework for potential development of protein binders.
Our recent structure of the complex of the human receptor:ligand interaction pair NKR-P1:LLT1 forms an excellent example. The human natural killer cell surface receptor NKR-P1 interacts with its cognate ligand LLT1 in an interesting manner, relying on two CTL:CTL interaction modes (Fig. 1) [2]. This interaction is one of the key components in recognition of tumour or virus-infected cells by immune system. The pair represents one of the CTL:CTL interactions with low affinity and several typical features of the “Canonical CTL:protein interaction” [1]. Comparison of the hNKR-P1:LLT1 interaction with other similar mammalian interaction pairs conforms to our structural classification of the CTL:CTL interactions.
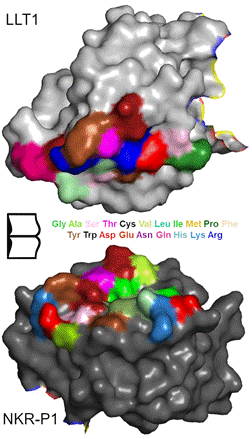
Figure 1. Primary interaction interface of human NKR-P1 and its ligand LLT1. An “open book” view with the interacting residues coloured according to amino acid type.
1. J. Dohnálek, T. Skálová, Biotechnol. Adv., 58, (2022), 107944.
2. J. Bláha, T. Skálová, B. Kalousková, O. Skořepa, D. Cmunt, V. Grobárová, S. Pazicky, E. Poláchová, C. Abreu, J. Stránský, T. Kovaľ, J. Dušková, Y. Zhao, K. Harlos, J. Hašek, J. Dohnálek, O. Vaněk, Nat. Commun., 13, (2022), 5022.
This work was supported by the Czech Science Foundation (18-10687S), by the ERDF fund (CZ.02.1.01/0.0/0.0/16_013/0001776 and CZ.02.1.01/0.0/0.0/15_003/0000447), and by the Czech Academy of Sciences (86652036).