Atomistic computer simulations of the ribosome
M. H. Kolář
Department of Physical Chemistry, University of Chemistry and Technology, Technická 5, 166 28 Prague
kolarh@vscht.cz
Atomistic computer simulations have become a valuable tool for understanding the behavior of biological macromolecules at the atomic level. We focus on the ribosome, one of the most complex and essential cellular machines, which is responsible for protein synthesis in all known organisms. The ribosome simulations are especially helpful in addressing questions about conformational heterogeneity and energetics of various ribosome parts. This way the simulations complement other biophysical techniques like cryogenic electron microscopy or fluorescent labeling [1, 2]. Despite the progress in hardware and software development, atomistic simulations of the ribosome remain challenging. The main reason is the ribosome size and complex chemical compositions. Still, using world-class supercomputers, one may gather valuable data in reasonable time.
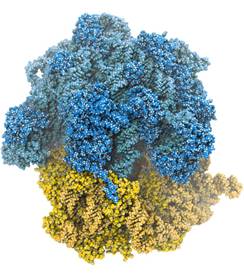
Figure 1: A bacterial ribosome with the small subunit in orange and large subunit in blue. Darker colours represent ribosomal proteins, ligher is the rRNA. Hydrogen in blue. Adopted from Ref. 2.
Over the past few years, we have been using molecular dynamics (MD) simulations to investigate several ribosome’s critical sites. Namely, we have studied the exit tunnel through which nascent proteins leave the ribosome [3], the decoding center where correct tRNAs are recognized or a portion of ribosome surface where translation factors bind [4]. In the talk, we will highlight the results of these projects and discuss how effective the computer simulations are in approaching scientific questions about biomolecules.
1. L. V. Bock, M. H. Kolář, H. Grubmüller, Curr. Opin. Struct. Biol., 49 (2018), 27-35.
2. L. V. Bock, S. Gabrielli, M. H. Kolář, H. Grubmüller, Ann. Rev. Biophys., 52 (2023), 0000.
3. M. H. Kolář, G. Nagy, J. Kunkel, S. A. Vaiana, L. V. Bock, H. Grubmüller, Nucleic Acids Res., 50, (2022), 2258-2269.
4. H. McGrath, M. Černeková, M. H. Kolář, Biophys. J., 121, (2022), 4443-4451.
The research was supported by the Czech Science Foundation (projects 19-06479Y and 23-05557S).