Structure and biological functions of TBEV Capsid protein
M. Selinger1,2, R. Novotný3,4, J. Sýs3,5, J. A. Roby6, H. Tykalová1, G. S. Ranjani7, M. Vancová2, F. Kaufman3, M. E. Bloom8, Z. Zdráhal7, L. Grubhoffer1,2, J. K. Forwood6, R. Hrabal4, M. Rumlová3 and J. Štěrba1
1Faculty of Science, the University of South Bohemia in České Budějovice, 370 05 České Budějovice, Czech Republic
2Institute of Parasitology, Biology Centre of the Czech Academy of Sciences, 37005 České Budějovice, Czech Republic
3Department of Biotechnology, University of Chemistry and Technology Prague, 16628 Prague, Czech Republic
4Laboratory of NMR Spectroscopy, University of Chemistry and Technology Prague, 16628 Prague, Czech Republic
5Institute of Organic Chemistry and Biochemistry, Czech Academy of Sciences, 16000 Prague, Czech Republic
6School of Dentistry and Medical Science, Charles Sturt University, NSW, Australia, 2678
7Central European Institute of Technology (CEITEC), Masaryk University, 62500 Brno, Czech Republic
8Biology of Vector-Borne Viruses Section, Laboratory of Virology, Rocky Mountain Laboratories, NIAID/NIH, 59840 Hamilton, MT, USA
Radim3.Novotny@gmail.com
Tick-borne encephalitis virus (TBEV) is the most medically relevant tick-transmitted flavivirus in Eurasia, targeting the host central nervous system and frequently causing severe encephalitis [1]. The primary function of its capsid protein (TBEVC) is to recruit viral RNA and form nucleocapsid [2].
Here we show the first 3D structure of a member of tick-born flavivirus C protein, TBEVC. The structure of monomeric D16-TBEVC was determined using high-resolution multidimensional NMR spectroscopy. Based on natural in vitro TBEVC homodimerization, the dimeric-interfaces were identified by hydrogen deuterium exchange mass spectrometry (Fig. 1). Although assembly of flaviviruses takes place in endoplasmic reticulum-derived vesicles [2], we observed that TBEVC protein also accumulated in nuclei and nucleoli of infected cells. Predicted bipartite nuclear localization sequence (bNLS) in the TBEVC C-terminal part was confirmed experimentally. The interface between TBEVC bNLS and import adapter protein importin-alpha was described using X-ray crystallography. Co-immunoprecipitation coupled with mass spectroscopy identification revealed 214 interaction partners of TBEV C including viral E and NS5 proteins and a wide variety of proteins involved mainly in rRNA processing and translational initiation.
Described findings may substantially help to design a targeted therapy against TBEV based on the novel functions of its capsid protein.
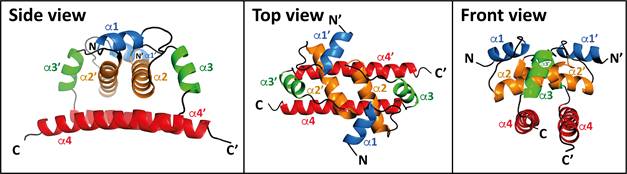
Figure 1: NMR structure of D16-TBEVC homodimer represented as cartoon. The monomeric protein consists of four a-helices marked as a1-a4. Generated with the PyMOL program (Schrödinger).
1. Gould EA, Solomon T: Pathogenic flaviviruses. Lancet (London, England) 2008, 371(9611):500-509.
2. Barrows NJ, Campos RK, Liao KC, Prasanth KR, Soto-Acosta R, Yeh SC, Schott-Lerner G, Pompon J, Sessions OM, Bradrick SS et al: Biochemistry and Molecular Biology of Flaviviruses. Chemical reviews 2018, 118(8):4448-4482.
We thank Z.Vavrušková (Institute of Parasitology, Biology Centre, CAS, Czech Republic) for the assistance in derivation of antibodies.
Computational resources were supplied by the project "e-Infrastruktura CZ" (e-INFRA CZ LM2018140 ) supported by the Ministry of Education, Youth and Sports of the Czech Republic.
The work was supported from European Regional Development Fund; OP RDE; Project: "ChemBioDrug" (No. CZ.02.1.01/0.0/0.0/16_019/0000729).