Zinc-dependent nuclease from Stenotrophomonas maltophilia: Structural analysis of the active site
K. Adámková1, 2, T. Kovaž1, M. Trundová1, B. Husáková1,2, J. Dohnálek1
1Institute of Biotechnology of the Czech Academy of Sciences, v.v.i., Průmyslová 595, 252 50 Vestec, Czech Republic
2University of Chemistry and Technology Prague, Department of Biochemistry and Microbiology, Technická 5, 166 28 Prague 6, Czech Republic
adamkovak@ibt.cas.cz
Zinc-dependent nucleases from S1-P1 family are relatively small globular proteins composed mostly of α‑helices, stabilized by disulfide bridges, and in the case of eukaryotic members they are also glycosylated. Gene coding an S1-P1 type nuclease (~25 % of sequence identity) can be found in many species, e.g. in plants, fungi, protozoan parasites, and also in some bacteria. Despite very similar active site composition and binding possibilities, these nucleases are able to cleave RNA, and single-stranded DNA as well as double-stranded DNA with different catalytic efficiency and substrate preferences [1].
The subject of our study is recombinant class I nuclease from Stenotrophomonas maltophilia (SmNuc1). Stenotrophomonas maltophilia is a Gram-negative aerobic bacterium from Gammaproteobacteria. It is an opportunistic human pathogen with a multiple antibiotic and stress resistance, infecting primarily severely immunocompromised patients and causing several nosocomial diseases [2]. Here we present novel structure of native SmNuc1 nuclease obtained at 1.4 Å resolution, followed by structures of complexes with 5′‑mononucleotides and structures of mutants of SmNuc1.
Analysis of these high-resolution crystal structures revealed several interesting novel features. Near the active site, there is a flexible loop able to open and close, which brings up new questions about the catalytic mechanism. Our structure-activity study of SmNuc1 and its variants could shed light on some aspects of the cleavage mechanism of this whole family of nucleases.
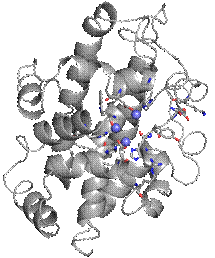
Figure 1: Crystal structure of SmNuc1 nuclease. Residues of the active site are shown as sticks and zinc ions are shown as blue spheres. Graphics was created using PyMOL (Schrödinger).
1. Kovaž T, Dohnálek J, Biotechnology Advances, 2018, 36(3): 603-612
2. Brooke J. S., Clinical Microbiology Reviews, 2012, 25(1): 2-41
The work was supported by the institutional support of IBT CAS, v.v.i. (RVO: 86652036), ERDF (CZ.02.1.01/0.0/0.0/15_003/0000447, CZ.02.1.01/0.0/0.0/16_013/0001776 and CZ.1.05/1.1.00/02.0109), MEYS CR (CZ.02.1.01/0.0/0.0/16_019/0000778 and LM2018127, support of Biocev-CMS core facilities Crystallization of Proteins and Nucleic Acids, and Structural Mass Spectrometry of CIISB, part of Instruct-ERIC) and by specific university research (grant No A1_FPBT_2021_003).