Biophysical characterization of novel monoclonal antibodies targeting epitopes on the SARS-CoV-2 Spike protein
O. Cehlar1, R. Skrabana1,2, K. Tomkova1,2, A. Kovac1,2, P. Filipcik1,2, N. Turic Csokova1, E. Kontsekova1,2, N. Zilka1,2, B. Kovacech1,2,3
1Institute of Neuroimmunology, Slovak Academy of Sciences; Bratislava, 845 10, Slovakia
2Axon Neuroscience R&D Services SE; Bratislava, 811 02, Slovakia
3 Axon Covidax a.s.; Bratislava, 811 02, Slovakia
ondrej.cehlar@savba.sk, kovacech@axon-neuroscience.eu
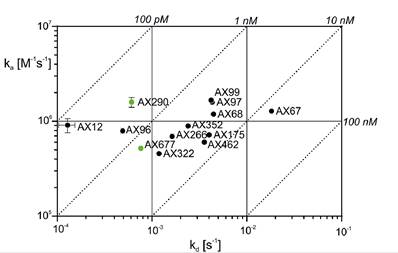
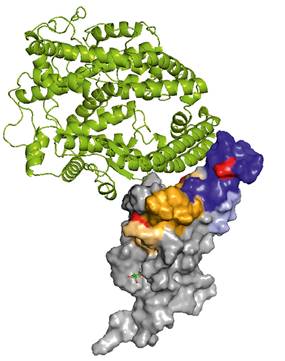
Clinical trials suggest that antibody treatments can prevent deaths and hospitalizations among people with mild or moderate COVID‑19. Considering high rate of SARS-CoV-2 escape mutations, a development of universal neutralization antibodies able to neutralize current and new variants of the virus is of a great importance. Employing hybridoma technology, a panel of anti SARS-CoV-2 Spike protein antibodies with a high affinity to the Spike receptor binding domain (RBD) has been generated (Fig. 1). Antibodies were selected based on the results of authentic virus neutralization assay. The antibodies AX290 and AX677, with non-overlapping epitopes on the Spike RBD (Fig. 2), showed excellent neutralization of an authentic SARS-CoV-2 virus representing strains circulating in Europe in spring 2020 and also the variants of concern B.1.1.7 (Alpha), B.1.351 (Beta) and B.1.617.2 (Delta). Unlike the majority of currently available therapeutic antibodies, AX677 is able to bind Omicron Spike protein just like the wild type Spike [1].
We have crystallized several antibodies alone and in the complex with RBD and characterize them on a synchrotron X-ray source.
|
|
|
Figure 1. Kinetic characteristics of the interactions of RBD with selected neutralizing antibodies represented with isoaffinity lines (dotted) of the individual ka, kd and KD affinity constants. The figure was adapted from ref [1]. |
Figure 2. The positions of the peptides bound by the antibodies, highlighted in the structure of SARS-CoV-2 spike RBD bound to ACE2 (PDB ID 6M0J [2]). The AX290 binding site is represented by shades of blue, AX677 by orange and yellow. ACE2 is shown in a green cartoon model, RBD as a grey surface model. The figure was adapted from ref [1]. |
1. Kovacech, B., Fialova, L., Filipcik, P., Skrabana, R., Zilkova, M., Paulenka-Ivanovova, N., Kovac, A., Palova, D., Rolkova, G.P., Tomkova, K., Csokova, N.T., Markova, K., Skrabanova, M., Sinska, K., Basheer, N., Majerova, P., Hanes, J., Parrak, V., Prcina, M., Cehlar, O., Cente, M., Piestansky, J., Fresser, M., Novak, M., Slavikova, M., Borsova, K., Cabanova, V., Brejova, B., Vinař, T., Nosek, J., Klempa, B., Eyer, L., Hönig, V., Palus, M., Ruzek, D., Vyhlidalova, T., Strakova, P., Mrazkova, B., Zudova, D., Koubkova, G., Novosadova, V., Prochazka, J., Sedlacek, R., Zilka, N., Kontsekova, E. eBioMedicine 76, (2022), 1–24.
2. Lan, J., Ge, J., Yu, J., Shan, S., Zhou, H., Fan, S., Zhang, Q., Shi, X., Wang, Q., Zhang, L., Wang, X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581, (2020), 215–220.
The study was funded by AXON Neuroscience SE and AXON COVIDAX a.s. The synchrotron data was collected at beamline P13 operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany). We would like to thank I. Bento for the assistance in using the beamline and to J. Stransky for crystal characterization at Centre of Molecular Structure, BIOCEV, Czech Republic