Multiapproach docking study for binding of intrinsically disordered tau peptides to monoclonal antibodies
S. Njemoga1,2, K. Meskova1, O. Cehlar1
1Institute of Neuroimmunology, Slovak Academy of Sciences, Dubravska cesta 9, 845 10 Bratislava, Slovakia
2Faculty of Natural Sciences, Comenius University, Ilkovicova 3278/6,
841 04 Bratislava, Slovakia
njemoga2@uniba.sk
Microtubule associated protein tau is the main actor of tau hypothesis of Alzheimer´s disease origin [1]. Under pathological conditions, by hyperphosphorylation of amino acid residues of the tau polypeptide chain, or upon truncation of tau protein, tau dissociates from microtubules. At the same time disintegration of microtubule and aggregation of tau monomers occur. Both of these events subsequently lead to nerve cell damage. Although it has not yet been confirmed whether this process is the trigger for Alzheimer's disease, the presence of insoluble aggregates of tau has been shown to be a hallmark of AD [2]. Therefore, tau protein appears to be a potential molecular target in the AD cure search process, whereas the main idea would be to design an inhibitor of tau aggregation to prevent disease progression. But we must first resolve the structural features of tau protein. Because of its intrinsically disordered character, tau doesn´t acquire any stable secondary structure. Monoclonal antibodies appear to be a useful tool to address the structural issue of tau, mainly their use as crystallization chaperones or in our case as docking and molecular simulation partners [3,5]. In addition to crystallization of tau complex with monoclonal antibody, in silico methods were implemented to obtain the transient tau structure.
Monoclonal antibody DC11 discriminates very strictly between physiological tau proteins and truncated tau peptides. This indicates the presence of a conformational epitope which carries the pathological function of tau and which is recognized by the mentioned antibody [3,4]. Therefore, we focused on the crystallization of the Fab fragment of DC11 antibody with tau321-391 to determine the conformational epitope of tau and to approximate the transition of tau from physiological to pathological conformation. Crystals of DC11Fab alone gave diffraction up to 1.4 Å. We also plan to dock the most populated clusters of tau conformations observed during MD simulations into the DC11 apo Fab structure.
A pan-tau monoclonal antibody DC25 recognizes tau epitope Lys347-Lys353 [5]. Using FlexPepDock and CABS-dock webservers we performed docking of flexible peptide tau347-353 into DC25 antibody structure with modest flexibility in CDR regions. From the resulting modelled complexes we have observed also the helical propensity of tau which was proposed also by the FELLS predictor.
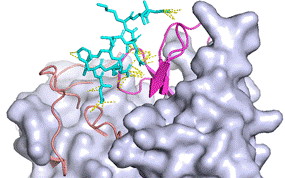
Figure 1: Docking result of tau347-353 with DC25 Fab from CABS-dock. The CDR regions of DC25 light chain are indicated as salmon cartoon and heavy chain CDR regions as magenta cartoon, tau is shown as cyan sticks, Fab framework regions as grey surface and contacts between chains within 3.0 Å are indicated in yellow (http://biocomp.chem.uw.edu.pl/CABSdock).
1. P. P. Liu, Y. Xie, X.Y. Meng, J.S. Kang, Signal Transduction and Targeted Therapy 4, (2019).
2. H. Braak, D. R. Thal, E. Ghebremedhin, K. Del Tredici, Journal of Neuropathology and Experimental Neurology 70, (2011), 960–969.
3. L. Vechterova, E. Kontsekova, N. Zilka, M. Ferencik, R. Ravid, M. Novak, NeuroReport 14, (2003), 87–91.
4. B. Kovacech, M. Novak, Current Alzheimer Research 7, (2010), 708–716.
5. O. Cehlar, R. Skrabana, A. Kovac, B. Kovacech, M. Novak, Acta Crystallographica Section F: Structural Biology and Crystallization Communications 68, (2012), 1181–1185.
This work was supported by the Scientific Grant Agency of the Ministry of Education of the Slovak Republic (grant no. VEGA 2/0145/19). The synchrotron data was collected at P13 beamline operated by EBML Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany). We would like to thank I. Bento fot the assistance in using the beamline.