Fragment-based characterization of substrate for novel FAD-dependent oxidoreductase from Chaetomium thermophilum
L. Švecová1, T. Skálová1, T. Kovaľ1, L. H. Østergaard2, J. Dohnálek1
1Institute of Biotechnology of the Czech Academy of Sciences, v.v.i., Průmyslová 595, Vestec, 252 50, Czech Republic
2Novozymes A/S, Brudelysvej 26, DK-2880 Bagsværd, Denmark
leona.svecova@ibt.cas.cz
Novel FAD-dependent oxidoreductase from lignocellulose-degrading fungus Chaetomium thermophilum (CtFDO) has a potential for industrial fields processing the lignocellulosic biomass, the most abundant material in the world. CtFDO is a member of glucose-methanol-choline (GMC) superfamily of oxidoreductases which act on hydroxyl groups of non-activated alcohols, carbohydrates and sterols. They share a two-domain character, conserved FAD-binding GxGxxG motif, and usually His-His or His-Asn catalytic pair in the active site accessible via a narrow tunnel or covered by a flexible loop [1].
The CtFDO crystal structure reveals a unique His-Ser active-site pair in a large wide-open pocket further extended beyond the pyrimidine moiety of FAD. These features indicate a different type of substrate than what is common for GMC oxidoreductases. This was confirmed by CtFDO enzymatic activity tests which, however, also excluded small lignin moieties and about 950 other compounds. To define the nature of the substrate, the crystallographic fragment-screening approach has been utilized. A series of six complexes led to identification of five binding subsites inside the active-site pocket with high preference for binding of aromatic moieties (Fig. 1). The conformational flexibility of interacting amino acids allows binding of compounds with a molecular weight greater than 500 Da. Our results indicate a complex polyaromatic nature of putative substrate, possibly with the character of larger lignin-building units [2] (Fig. 1).
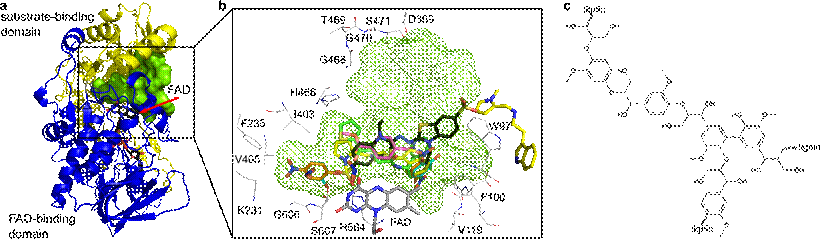
Figure 1 (a) Crystal structure of CtFDO with color-coded substrate-binding (yellow) and the FAD-binding (blue) domains. FAD is shown as sticks with black C atoms and the active-site pocket with green surface. (b) Three-dimensional superposition of active sites of the CtFDO complexes. Residues surrounding the pocket (green mesh) and FAD are shown as sticks with gray C atoms and the ligands with orange, cyan, black, yellow, green, and magenta C atoms. Graphics were created using PyMOL (Schrödinger). (c) Chemical structure of lignin.
1. L. Sützl, G. Foley, E. M. J. Gillam, M. Bodén, D. Haltrich, Biotechnol Biofuels, 12, (2019), 1–18.
2. L. Švecová, L. H. Østergaard, T. Skálová, K. Schnorr, T. Koval’ et al., Acta Cryst., D77, (2021), 755–775.
The work was supported by the institutional support of IBT CAS, v.v.i. (RVO: 86652036), ERDF (CZ.02.1.01/0.0/0.0/15_003/0000447, CZ.02.1.01/0.0/0.0/16_013/0001776 and CZ.1.05/1.1.00/02.0109), MEYS CR (CZ.02.1.01/0.0/0.0/16_019/0000778 and LM2018127, support of Biocev-CMS – core facilities Biophysical Methods, Crystallization of Proteins and Nucleic Acids, and Structural Mass Spectrometry of CIISB, part of Instruct-ERIC) and by the GA CTU in Prague (SGS19/189/ OHK4/3T/14).