Cryo-EM ensemble reveals the mechanism of +1 ribosomal frame shifting
G. Demo1,2, H.B. Gamper3, A.B.
Loveland2, I. Masuda3, C.E. Carbone2,
E. Svidritskiy2, Y.M. Hou3, A.A. Korostelev2
1Central European Institute of Technology, Masaryk University, Brno, Czech Republic
2RNA Therapeutics Institute, Department of Biochemistry and Molecular Pharmacology, UMass Medical School, Worcester, MA, USA
3Department of Biochemistry and Molecular Biology, Thomas Jefferson University, Philadelphia, PA, USA
gabriel.demo@ceitec.muni.cz
To accurately synthesize a protein, the ribosome maintains the mRNA reading frame by decoding and translocating one codon at a time [1]. Change of the reading frame of mRNA during translation, termed frame shifting, provides a strategy to expand the coding repertoire of cells and viruses [2]. The translating ribosome switches to an alternative reading frame, either in the forward (+) or reverse (–) direction, i.e., skipping or re-reading one or more mRNA nucleotides, respectively. For example, +1 frame shifting (+1FS) controls the expression of the essential release factor 2 in bacteria [3] and leads to pathological expression of huntingtin in eukaryotes [4]. How and where in the elongation cycle +1FS occurs remains poorly understood.
Here we address this challenge by using cryo-EM to visualize +1FS on +1FS-prone mRNA sequences. We present cryo-EM structures of 70S complexes, allowing visualization of elongation and translocation by the GTPase elongation factor G (EF-G). Four structures with a +1FS-prone mRNA reveal that frame shifting takes place during translocation of tRNA and mRNA. The +1FS-prone pre-translocation complex maintains the 0-frame anticodon–codon pairing resembling that in canonical elongation complexes. In the mid-translocation complex with EF-G, the tRNA shifts to the +1-frame near the P site, with bulged nucleotide between the E and P-site codons stabilized by G926 on the 16S rRNA. The ribosome remains frame shifted in the nearly post-translocation state. Our findings reveal that the ribosome is pre-disposed for +1FS before translocation, and that frame shifting is accomplished at an intermediate stage of EF-G catalysed translocation (Fig. 1).
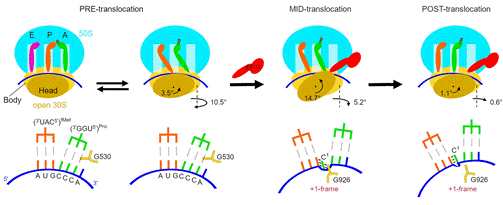
Figure 1. Schematic of ribosomal translocation by EF-G resulting in +1 frame shifting. The second row shows local rearrangements of mRNA-tRNA and positions of the decoding-center nucleotide G530 and P-site nucleotide G926 of the 30S subunit.
1. H. F. Noller, L. Lancaster, J. Zhou, S. Mohan. Nat. Struct. Mol. Biol., 24, (2017), 1021–1027.
2. J. D. Dinman. Adv. Protein Chem. Struct. Biol., 86, (2012), 129–149.
3. W. J. Craigen, C. T. Caskey. Nature, 322, (1986), 273–275.
4. P. Saffert, F. Adamla, R. Schieweck, J. F. Atkins, Z. Ignatova. J. Biol. Chem., 291, (2016), 18505–18513.
This study was supported and funded by Czech Science Foundation, project no. GJ20-16013Y (to G.D.)