Mechanism of replicative "rolling-circle" DNA transposition in eukaryotes
D. Kosek1,5, I. Grabundzija1,2, H. Lei1, I. Bilic2, H. Wang3, Y. Jin4, G.F. Peaslee4, A.B. Hickman1, F. Dyda1
1 Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892, USA
2 BioNTech Cell & Gene Therapies GmbH, 55131 Mainz, Germany
3 Multi-Institute Cryo-Electron Microscopy Facility, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892, USA
4 Department of Physics, University of Notre Dame, Notre Dame, IN 46556, USA
5 Department of Structural Biology of Signaling Proteins, Institute of Physiology, CAS, BIOCEV, Prumyslova 595, 252 50 Vestec, Czech Republic
dalibor.kosek@fgu.cas.cz
Evolution is driven forward by the acquisition of novel traits which rise as a consequence of changes in the heritable genome. There are many ways how changes in DNA could occur, including recombination, replication errors, inaccurate repair or chemical modifications. One of the most important drivers of such changes is transposition, the protein-catalyzed transfer of discrete mobile DNA elements from one site into another. These „selfish” DNA elements (transposons) often comprise a vast amount of non-coding genome in eukaryotic species and have contributed to their evolution in many ways. Their movement not only disrupts and reorders DNA but it can also provide regulatory sequences that enable the establishment of novel expression patterns providing an evolutionary framework for rapid adaptation in unfavorable environment[1].
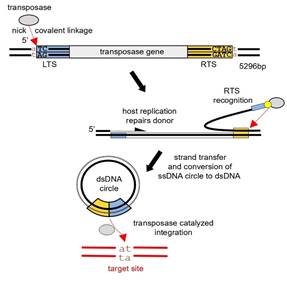
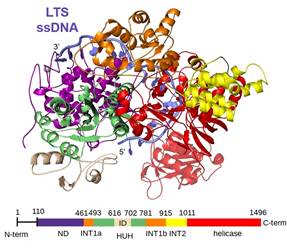
Helitrons are ancient DNA transposons that have dramatically reshaped many eukaryotic genomes due to their large numbers and propensity to capture and mobilize host gene fragments. They have contributed to genome diversity with their unusual replicative transposition mechanism that, unlike the better-characterized cut-and-paste DNA transposons, relies on ssDNA intermediates. The current model for the Helitron transposition mechanism suggests that the initial nicking at the left transposon end generates free 3’-OH group which primes the DNA replication displacing the leading transposon DNA strand. When the other end is reached, a second cleavage reaction and strand transfer generate an excised circular Helitron intermediate. This can then be integrated at a new genomic location (Figure 1). Here, we describe the cryo-EM structure of the monomeric transposase from the recently reconstituted active Helitron element[2] covalently bound to the 5’-end of the transposon ssDNA (Figure 2). This represents the first structural view into the molecular architecture and function of Helitron transposases, the largest transposases known to date. It reveals how Helitrons have solved the problem of uncoordinated reactions on its two transposon ends: the transposase protects the initially cleaved and displaced end by forming a tightly packed assembly that buries covalently bound ssDNA in the core of HUH nuclease and Pif1-like helicase domains with a scaffold formed by other surrounding domains, some with apparently unique protein folds. Despite the lack of evidence for a direct evolutionary link, the architecture is reminiscent of that seen in TraI, a prokaryotic relaxase involved in bacterial horizontal gene transfer. Our work also suggests the structural basis of the interplay between two juxtaposed active site tyrosines that alternate in the roles of the catalytic nucleophiles in the HUH active site; this result is likely generalizable to all proteins that use an HUH Y2 nucleases to initiate replication[3]. We have combined the structural work with in vitro biochemical studies that probe the role of the two active site tyrosines as well as in vivo transposition assays that suggested different roles of each tyrosines during the transposition.
This work presents the first three-dimensional insight into a large and important superfamily of eukaryotic transposases [4]. The structure breaks the paradigm that DNA transposases must function as multimers, thus, expanding the conceptual framework for understanding the mechanisms of transpositional DNA rearrangements. It also represents the first structural insight into the coupled actions of an HUH-type nuclease and a helicase – poorly understood arrangement widespread not only in relation to the transposition but also viral replication and propagation of antibiotic resistances. Also, in principle a monomeric transposase opens up possibilities to novel genome targeting strategies simpler than those that have been recently attempted with multimeric transposase systems.
|
|
||
|
|
|
|
Figure 1. Cartoon schematic of the reconstituted Helitron transposon with current model for Helitron transposition. Helitron is bordered by 150 bp of LTS (left terminal sequence; in blue throughout) and 150 bp of RTS (right terminal sequence; in yellow throughout). Red arrows indicate cleavage or strand transfer reactions. Yellow circle marks a covalent linkage. |
Figure 2. Cartoon view of cryo-EM determined structure of Helitron transposase in complex with ssDNA of LTS (in blue) with domains colored as indicated in the schematic below. ND, N-terminal domain; INT1, intermediate domain 1; HUH, catalytic HUH domain with insertion (ID); INT2, intermediate domain 2. |
|