NACHRDB: solving the puzzle of structure-function relationships in nicotinic acetylcholine receptors (nAChRs)
A. Chareshneu1,2*, P. Pant1,2, R. J. T. Ramos1,2, T, Gokbel C.-M.3, Ionescu1,2, J. Koča1,2
1CEITEC - Central European Institute of Technology, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic
2National Centre for Biomolecular Research, Faculty of Science, Masaryk University, Kamenice 5, 625 00 Brno, Czech Republic
3Department of Molecular Biology and Genetics, Izmir Institute of Technology, Izmir, Turkey
479052@mail.muni.cz
The nicotinic acetylcholine receptor (nAChR) is an evolutionary ancient large (~300 kDa) allosteric membrane protein mediating the synaptic transmission [1]. This prototypic member of the family of pentameric ligand-gated ion-channels is involved in many physiological processes (from learning to motor control), neurological diseases (Alzheimer’s and Parkinson’s diseases, schizophrenia, epilepsy), and addictions (alcohol, tobacco) [2]. Since its biochemical isolation in 1970, extensive studies generated huge amounts of structural-functional data. However, the cumulative knowledge on nAChRs, spanning ~50 years of research, is not systematically accessible. The wide variety in receptor types, residue numbering schemes, and methods used, together with diverse terminology, the absence of comprehensive structural annotation, and the scattered nature of the existing findings make it harder to summarize the current knowledge and apply it efficiently to promote further discoveries. There is no single resource providing an access to and visualization of such diverse, complex, and extensive information. To fill this gap, we developed NACHRDB (https://crocodile.ncbr.muni.cz/Apps/NAChRDB/) – a web-accessible manually curated database which not only provides intuitive and fast access to relevant structural-functional data on nAChRs, but also facilitates its interpretation by integrating the residue-level annotations with interactive and highly responsive visualization of sequence and 3D structure [3]. Besides, NACHRDB can provide the users with a prediction of residues potentially relevant for the allosteric regulation of nAChRs, based on the analysis of partial atomic charges profile. We believe that NACHRDB not only can guide the further studies in the field of nAChRs, helping the researchers to detect hitherto unknown association between structure and function of nAChRs, but also serve as a key starting point in unification of the state-of-art knowledge in the broad field of ion channels.
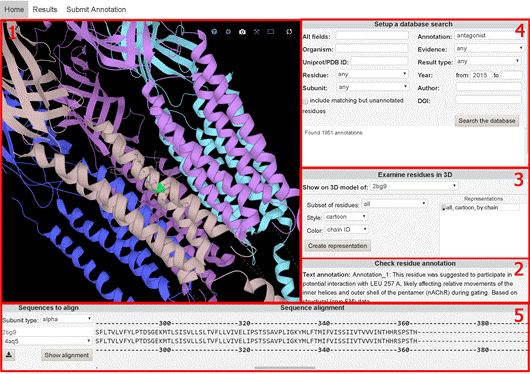
Figure 1. The NAChRDB workspace. The Home tab contains: (1) an interactive 3D visualization of nAChR model, (2) annotation records for a selected residue, (3) 3D visualisation settings, (4) a search section for querying structural, functional, and literature-related information; (5) a sequence alignment viewer. The Results tab (B) provides the search results in the form of an interactive sorted table. All results are available for download in. csv and. json format. The Submit tab (C) provides an opportunity for the users to report annotation information, contributing to NAChRDB data maintenance.
1. Changeux J-P. The nicotinic acetylcholine receptor: a typical “allosteric machine”. Philos Trans R Soc Lond B Biol Sci. 2018;373. doi:10.1098/rstb.2017.0174
2. Albuquerque EX, Pereira EFR, Alkondon M, Rogers SW. Mammalian Nicotinic Acetylcholine Receptors: From Structure to Function. Physiol Rev. 2009;89: 73–120. doi:10.1152/physrev.00015.2008
3. Chareshneu A, Pant P, Ramos RJT, Gökbel T, Ionescu C-M, Koča J. NAChRDB: A Web Resource of Structure-Function Annotations to Unravel the Allostery of Nicotinic Acetylcholine Receptors. bioRxiv. Cold Spring Harbor Laboratory; 2020; 2020.01.08.898171. doi:10.1101/2020.01.08.898171
We would like to acknowledge Dr. David Sehnal for his help with the implementation of 3D viewer based on the LiteMol suite.