Preliminary characterization of human CD160 and the influence of viral N-linked glycosylation related to this immune inhibitory pathway
S. Lenhartová, V. Kempová, M. Benko, M. Nemčovič, I. Nemčovičová
Biomedical Research Center, Slovak Academy of Sciences, Bratislava
simona.lenhartova@savba.sk
The CD160 (BY55) receptor was recently identified as a candidate target for immmunotherapy. Its expression is mainly, but not uniquely, restricted to cytotoxic cells, such as NKT cells and NK cells, and it is also present in T cells [1]. The relationship of human CD160 to HVEM (TNF receptor family member 14), and its viral ortholog UL144 (encoded by human cytomegalovirus, HCMV) are nowadays being extensively studied because of their specific properties in immune modulation. However, molecular characterization and binding properties of these molecules are not clear yet. Our biophysical and molecular characterization results provide insight into the basis for understanding of mechanisms by which CD160 modulates immune effector pathways that might be influenced by HCMV glycosylation.
Materials and methods
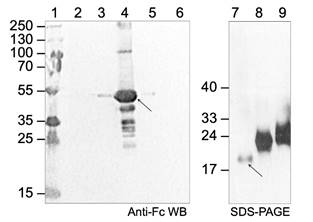
Construction of recombinant baculoviruses (bacmids) with target gene of CD160, was engineered by PCR and mature ectodomain of protein was cloned downstream of the gp67 signal sequence into the baculovirus transfer vector pAcGP67A containing an N-terminal Fc-tag. The recombinant bacmid with target gene was transfected into Sf9 cells to produce CD160 Fc-fused protein (about 52-60 kDa, Figure 1) via ProGreen™ Baculovirus DNA carrying a GFP reporter gene (AB Vector). After several rounds of viral amplification in Sf9 cells, the recombinant protein was purify after 3 days from the cultivation medium of cultured Sf9 cells infected with baculovirus according to previously reported conditions [2]. The purity, molecular mass, and the conformational changes of the protein were determined by various liquid chromatography (LC: FPLC), gel electrophoresis (SDS-PAGE) and western blot (WB) analysis (Figure 1). Kinetic binding studies of synthetized protein ectodomains were performed by surface plasmon resonance (SPR) equilibrium binding assays on BIACORE instrumentation and relative affinities were calculated. After a proper selection and establishment of the expression system followed by biochemical characterization at the protein level, we have started the initial screening for crystallization conditions in order to obtain suitable crystal for X-ray diffraction and structural studies.
 Figure 1. Western
blot analysis of CD160 Fc-tagged protein using the Anti-Human
IgG (Fc specific)-Peroxidase antibodies and SDS-PAGE analysis of HCMV
UL144-mutants.
Lane 1: PageRuler Plus Prestained Protein
Ladder
(Thermo
Scientific);
line 2: flow-through fraction, nonbinding proteins; lines 3-6: elution fractions;
lines 7-9: mutants of HCMV UL144 with different glycosylation. The arrows
indicate the CD160 Fc-tagged protein (52-60 kDa) and UL144-mutant (dg) used for
binding assay.
Figure 1. Western
blot analysis of CD160 Fc-tagged protein using the Anti-Human
IgG (Fc specific)-Peroxidase antibodies and SDS-PAGE analysis of HCMV
UL144-mutants.
Lane 1: PageRuler Plus Prestained Protein
Ladder
(Thermo
Scientific);
line 2: flow-through fraction, nonbinding proteins; lines 3-6: elution fractions;
lines 7-9: mutants of HCMV UL144 with different glycosylation. The arrows
indicate the CD160 Fc-tagged protein (52-60 kDa) and UL144-mutant (dg) used for
binding assay.
Results and Discussion
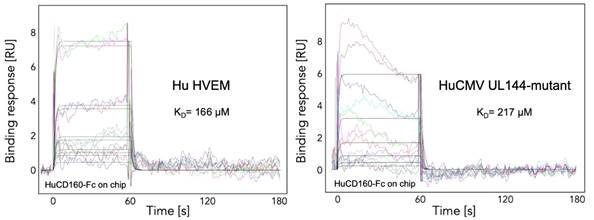
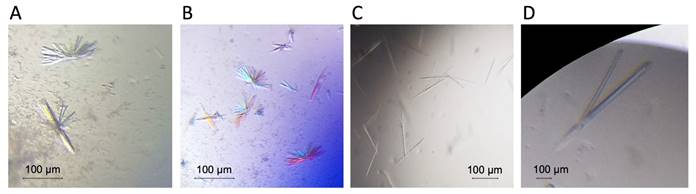
CD160 was identified as a co-inhibitory molecule that binds HVEM with a similar affinity to human checkpoint receptor BTLA [3]. Our binding studies of expressed ectodomains revealed slower dissociation rate for HVEM-CD160 than for HVEM-BTLA molecular complex. Our recent data (unpublished) suggest that HCMV UL144, viral mimic of the HVEM, does not bind CD160, we hypothesize this is due to altered N-linked glycosylation. Mutant of UL144 (dgUL144) lacking these glycosylation sites (N61, N70, N73, N78, N91, N99, N116) was generated and we have determined whether its binding to CD160 is impacted. We have measured the binding kinetics (by using Biacore SPR assay) for the dgUL144-CD160 (KD 217 mM) and HVEM-CD160 (KD 166 mM) molecular complexes (Figure 2). Additionally, we have performed the crystallization condition screening for CD160 alone (thrombin digested Fc-portion) and HVEM-CD160 complex. The preliminary crystallization conditions were found (Figure3) and will be further optimized and tested for X-ray diffraction that will provide critical insight into understanding of how CD160 function in regards to human HVEM and viral ortholog UL144 to regulate immune responses.
Figure 2. SPR analysis of HVEM-CD160 and dgUL144mutant-CD160 molecular complexes. Plots illustrating the experimental curve-fitting methodology for a simple binding model (1:1 Langmuir).
![]()
![]()
![]()
![]()
Figure 3. Preliminary crystallization of CD160-HVEM (A, B) complex and CD160 alone (C, D). Initial crystals were obtained under following conditions: 0.2 M ammonium chloride, 0.1 M HEPES pH 7.0, 20% PEG 6000 (Fig. 3 A, B); 0.2M sodium malonate dibasic monohydrate, 20 % PEG 3350 (Fig. 3 C); 0.2M sodium malonate dibasic monohydrate, 0.1 M Bis-Tris propane pH 6.5, 20 % PEG 3350 (Fig. 3 D).
References
- G. Cai, G. J. Freeman, Immunol. Rev., 229, (2009), 244.
2. I. Nemčovičová, C. A. Benedict, D. M. Zajonc, PLoS Pathogen, 9, (2013), e1003224.
3. R. Kojima, M. Kajikawa, M. Shiroishi, K. Kuroki, K. Maenaka, J. Mol. Biol., 413, (2011), 762-772.
Acknowledgements
The work was supported by the contribution of the Slovak Research and Development Agency under the project APVV-14-0839 and the contribution of the Scientific Grant Agency of the Slovak Republic under the grant 02/0020/18 and 02/0130/18. IN is Marie Curie Fellow financed by Programme SASPRO co-funded by European Union and the Slovak Academy of Sciences under the contract 0003/01/02. The part of the research team is supported by Interreg V-A SK-AT cooperation programme by project CAPSID under the contract No. NFP305010V235 co-financed by European Regional Development Fund.