Calcium Bridging between Carbohydrates and Lectins: A Difficult Case for Classical Molecular Dynamics
M. Lepšík, A. Imberty
Centre de recherches sur les macromolécules végétales (CERMAV), CNRS, Grenoble, France
martin.lepsik@cermav.cnrs.fr
The developments of additive carbohydrate force fields increased the reliability of molecular dynamics (MD) simulations of protein-carbohydrate complexes [1]. The presence of bridging Ca2+ ions can, however, pose problems for structural and energetic description due to quantum effects, such as polarisation and charge transfer [2, 3]. To overcome this limitation, we had developed Ca2+ parameters with effective electronic polarisation for use with additive force fields [2] and applied them to a calcium-dependent lectin/carbohydrate complex whose structure we have determined crystallographically (Fig. 1) [4]. Such a treatment improved the structural description of the binding site (Ca2+···Ca2+ distance) in submicrosecond MD but an extension to 1.4 µs showed instabilities in protein/carbohydrate interactions. Thus, a systematic testing of charge-group scaling and use of various water models was launched to determine a universal protocol to describe reliably lectin – calcium –carbohydrate complexes by MD. We propose that such protocols will be transferable to simulations of other charged biomolecular systems.
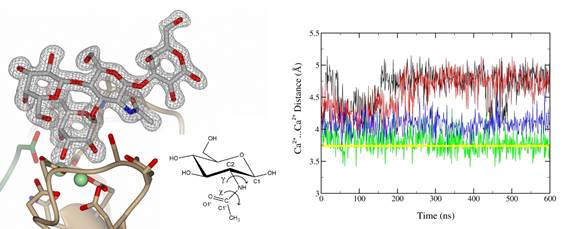
Figure 1. The binding mode of Lexis x tetrasaccharide complexed to two calcium ions in the binding site of LecB lectin of Pseudomonas aeruginosa. Dihedral angles γ and χ of N-acetyl glucosamine which are monitored during MD are shown. Ca2+···Ca2+ distances in several MD setups (classical Ca2+ parameters - black, red; scaled Ca2+ parameters – blue, green) are compared with the crystallographic value (yellow).
1. R. J. Woods, Chem. Rev., 118, (2018), 8005.
2. M. Kohagen, M. Lepsik, P. Jungwirth, J. Phys. Chem. Lett., 5, (2014), 3964.
3. M. Lepsik & M. J. Field, J. Phys. Chem. B, 111, (2007), 10012.
4. M. Lepsik, et al., Eur. J. Med. Chem., 177, (2019), 212.
ML acknowledges funding from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No. 795605, E.U. Computations were made possible thanks to the GRICAD infrastructure (https://gricad.univ-grenoble-alpes.fr), which is supported by Grenoble research communities, the HPC-EUROPA3 project (INFRAIA-2016-1-730897), E.U., with the support of the EC Research Innovation Action under the H2020 Programme, EPCC at the University of Edinburgh, Scotland and HPC resources from GENCI-IDRIS (Grant 2019-A0070711040).