Crystallography of fuzziness in the complexes of intrinsically disordered proteins
R. Skrabana1, R. Dvorsky2, O. Cehlar1, L. Fialova1, E. Kontsekova1
1Institute of Neuroimmunology, Slovak Academy of Sciences, Bratislava, Slovakia
2Institute of Biochemistry and Molecular Biology II, Heinrich-Heine University, Düsseldorf, Germany
Intrinsically disordered proteins (IDPs) and disordered protein regions have an intriguing property: their heterocomplexes with other cellular partners are often very stable, albeit having a large degree of structural heterogeneity (fuzziness, [1, 2]). The fuzziness is perceived as the inability to define a unique 3D-configuration of interacting interfaces. Prevalent methods for the fuzziness detection are ensemble and time averaging techniques like NMR, CD or single molecule fluorescence.
Studying complexes of IDP tau with antibodies by X-ray crystallography, we have noted that independently refined multiple copies of the complex in asymmetric unit may display contrasting details in their binding interfaces (Fig. 1). Crystallography thus may confer mostly unexpected contribution to the definition of binding contacts in fuzzy complexes, and, by implication, it may suggest the propensity of IDP to certain conformations already in the solution monomeric state.
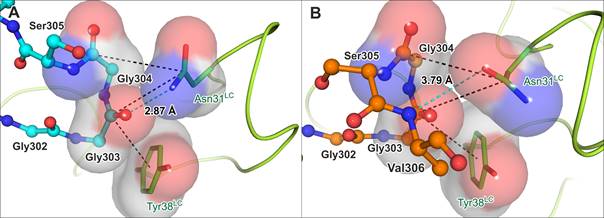
Figure 1. Different copies of antibody-tau complex in asymmetric unit exhibit contrasting conformations (PDB ID 5MP3).
(A) Tau chain C (cyan) in the Fab-p5 crystal structure. (B) Tau chain D (orange) in the Fab-p5 crystal structure. Hydrogen bonds are indicated by cyan dashes with the length in angstroms, π-interactions are in black dashes and π-electron interacting systems are visualized as space-filling models.