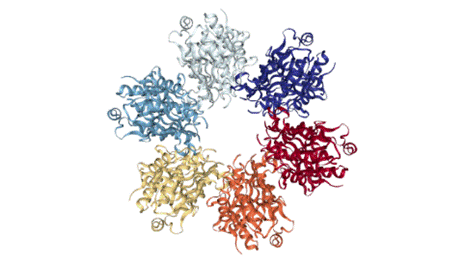
Fig. 1. The tertiary structure of P4 hexamer from
phage φ8. The monomers are distinguished by different colors [Data from
PDB: http://www.rcsb.org/3d-view/4BLQ/1].

The dsRNA bacteriophages
of Cystoviridae family package their
genome into empty capsid – procapsid, which protects the genome from degradation
inside as well as outside host cell. The genome packaging is performed by a molecular
motor - P4 proteins, which are components of procapsid. The P4s possess an NTPase
activity that converts the chemical energy from ATP hydrolysis to a mechanical
movement of packaging ssRNA precursors into a procapsid, where the replication
and transcription of dsRNA occurs [1- 3]. The P4s are RNA helicases belonging to the Superfamily
4 of helicases with characteristic presence of conserved sequence motifs (H1,
H1a, H2, H3 and H4) [1- 3]. The RNA helicases cause the distribution of RNA-protein complexes and
carry out RNA unwinding [2]. The P4 assembles into hexameric
ring (Fig.1), which has on the outer perimeter NTP-binding sites and the nucleic
acid binding sites are located in the central channel. Each P4monomer can be divided into N-terminal , core NTPase domain with
sequence motif and C-terminal domain, which is inserted into the central
channel of hexamer and its conformational changes regulate ring stability and ATPase
activity of P4s [1, 3-4]. Here we grow the monocrystals of the φ8 P4 protein (Fig. 2) in the crystallization conditions
of 100mM sodium acetate (pH 4,6) and 2,2 M ammonium sulphate. These conditions were
suitable enough to be a good starting point for the next crystallization
experiments with RNA assembled to P4.
The work is
supported from European Regional Development Fund-Project "Mechanisms and
dynamics of macromolecular complexes: from single molecules to cells" (No.
CZ.02.1.01/0.0/0.0/15_003/0000441).

