Characterization of a novel bidirectional NK activating ligand CD160
Ivana Nemčovičová1,3,, Marek Nemčovič2, Marcela Kúdelová1, Dirk Zajonc3
1Biomedical Research Center and 2Institute of Chemistry at the Slovak Academy of Sciences, Bratislava, Slovakia
3La Jolla Institute, Division of Cell Biology, La Jolla, California, USA
viruivka@savba.sk
Natural killer (NK) cells require cytokine signals to differentiate into fully functional effector cells that initiate protective antiviral responses [1]. Many pathogens have evolved countermeasures to avoid interferon-mediated detection and clearance by NK cells [1,2], however, regulatory mechanisms limiting cytokine activation of NK cells that reduce non-specific tissue damage remain poorly defined. CD160 is a 27 kDa glycoprotein which was initially identified with the monoclonal antibody BY55 [2]. Its expression is tightly associated with peripheral blood NK cells and CD8 T lymphocytes with cytolytic effector activity [2,3,4]. The cDNA sequence of CD160 predicts a cysteine-rich, glycosylphosphatidylinositol-(GPI)-anchored protein of 181 amino acids with a single Ig-like domain weakly homologous to KIR2DL4 molecule. It was found that TNF receptor herpesvirus entry mediator (HVEM) preferentially engages CD160 trimer to costimulate activation, while a viral ortholog of HVEM specifically binds to B and T lymphocyte attenuator (BTLA) to suppress this signaling. CD160 antigen is a protein that in humans is encoded by the CD160 gene. We have found that CD160 is expressed at the cell surface as a tightly disulfide-linked multimer. The homology model of CD160 antigen domain [Figure 1] shows cysteine-rich region that was found to be responsible for CD160 tight-timer formation even under reduced conditions. CD160 trimer forms stable complex with HVEM, while monomeric form refused to binds its cognate ligand. Thus, regulation of CD160 bidirectional binding may represent a common mechanism of immune regulation targeted by multiple pathogens, which by extension is a potential target for therapeutic manipulation.
Figure 1 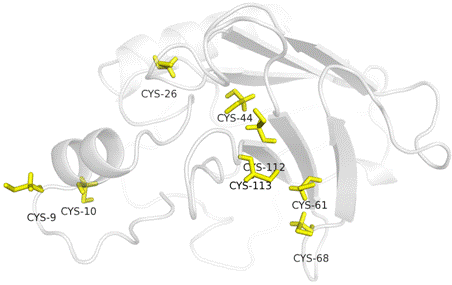
IN is Marie Curie Fellow financed by Programme SASPRO, co-funded by European Union and the Slovak Academy of Sciences. The authors gratefully acknowledge the contribution of the Slovak Research and Development Agency under the project APVV-14-0839 and the contribution of the Scientific Grant Agency of the Slovak Republic under the grant 2/0103/15.
1. Vivier E, et al., Science 2011, 331, 44-49
2. Anumanthan A, et al., J Immunol 1998, 161(6), 2780–90
3. Agrawal S, et al., J Immunol 1999, 162(3), 1223–26
4. Klopocki E, et al. Am. J. Hum. Genet. 2007, 80(2), 232–40