Lone-pair-p interactions in nucleic acids and proteins:
physical origin and significance
Jan Novotný,1 Sophia Bazzi,1,2,3 Radek Marek,1,3 Jiří Kozelka2,4
1. CEITEC – Central European Institute of Technology, Masaryk University, Kamenice 5/A4, CZ-625 00 Brno, Czech Republic
2. Department of Condensed Matter Physics, Faculty of Science, Masaryk University, Kotlářská 2, CZ-611 37 Brno, Czech Republic
3. National Center for Biomolecular Research, Faculty of Science, Masaryk University, Kamenice 5/A4, CZ-625 00 Brno, Czech Republic
4. Université Paris Descartes, UMR 8601 CNRS, 45, rue des Saints-Pères, 75270 Paris, France,
E-mail: kozelka.jiri@gmail.com
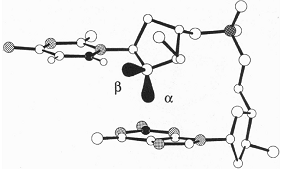
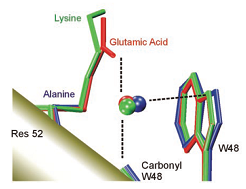
The lone-pair-p interaction is a still relatively unexplored bonding between a lone-pair (lp) of electrons of an electronegative atom and a p-system. In 1995, Egli and Gessner identified in the d(CpG) steps of Z-DNA an interaction between an oxygen lp of the cytidine deoxyribose and the guanine base, which they classified as n®p* hyperconjugation and related to the stability of Z-DNA [1]. A similar type of interaction was suggested to occur in many protein crystal sturctures where water molecules were observed to contact a tryptophan or a histidine residue along the normal to the ring plane through the endocyclic N atom [2].
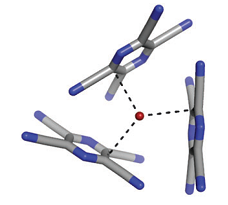
A subclass of lp-p interactions are anion-p interactions, where the interacting lp is a part of an anionic residue. For instance, electron-deficient arenes such as 1,2,4,5-tetracyanobenzene (TCB) or tetracyanopiperazine (TCP) form with halide ions in solution and in solid phase complexes whose charge-transfer absorption bands are indicative of a weakly covalent interaction. Since the halides interact with the p-face of the arene in a similar manner as does water with tryptophan in proteins and deoxyribose with nucleobases in nucleic acids (see figure below), on may ask to which extent charge transfer, evidently operating in the halide-arene complexes, contributes to the binding of the lp-p interactions in the biopolymers. Asking more generally: What are the energy components stabilizing lone-pair-p interactions and how does their balance depend on the molecular properties of the interacting partners? Do force-field calculations portray such interactions properly?
The talk will attempt to provide the answer.
|
|
|
|
Left: lp-p interaction stabilizing Z-DNA [1]. Middle: lp-p interaction suggested to operate between a conserved water molecule and tryptophan W48 in the Engrailed homeodomain and its mutants [2]. Right: lp-p interaction observed in the X-ray structure of the charge-transfer complex between Br- and TCP [3].
|
||
1. Egli, M. and Gessner, R., Proc. Nat. Acad. Sci. USA, 1995, 92, 180-184.
2. Stollar, E. J., Gelpi, J. L., Velankar, S., Golovin, A., Orozco, M. and Luisi, B. F., Proteins, 2004, 57, 1-8.
3. Rosokha, Y. S.; Lindeman, S. V.; Rosokha, S. V.; Kochi, J. K., Angew. Chem. Int. Ed., 2004, 43, 4650-4652.