Structural analysis of a novel haloalkane dehalogenase DbeA from Bradyrhizobium elkanii
USDA94
Tatyana Prudnikova1,2, Pavlina Rezacova3, Radka
Chaloupkova4, Yukari Sato5, Michal Kuty1,2, Tana Koudelakova4 Yuji Nagata5, Jiri Damborsky4, and Ivana
Kuta Smatanova1,2
1Faculty
of Science, University of South Bohemia
Ceske Budejovice, Branisovska 31, 370 05 Ceske Budejovice, Czech Republic
2Institute of Nanobiology and Structural Biology GCRC, Academy of Sciences of the Czech Republic, Zamek 136, 373 33 Nove Hrady, Czech Republic
3Institute of Organic Chemistry and Biochemistry and Institute of Molecular Genetics, Academy of Sciences of the Czech Republic, Flemingovo n.2, Prague 6, Czech Republic
4Loschmidt Laboratories, Department of Experimental Biology and Research Centre for Toxic Compounds in the Environment, Faculty of Science, Masaryk University, Kamenice 5/A4, 625 00 Brno, Czech Republic
5Department
of Environmental Life Sciences, Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira,
Sendai 980-8577, Japan
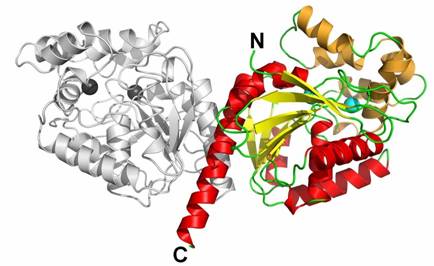
A novel enzyme, DbeA, belonging to the family of haloalkane dehalogenases (EC 3.8.1.5) was isolated from Bradyrhizobium elkanii USDA94. This haloalkane dehalogenase is closely related to DbjA enzyme from Bradyrhizobium japonicum USDA110 (71% sequence identity), but has different biochemical properties. DbeA is generally less active and has a higher specificity towards brominated and iodinated compounds than DbjA. The DbeA protein was crystallised using the sitting-drop vapour-diffusion method. The crystals of DbeA belong to the primitive orthorhombic space group P212121. Crystal structure of a DbeA enzyme has been solved and refined to 2.2 Å resolution. The enzymatic molecular structure of DbeA was compared with those of known haloalkane dehalogenases already deposited in Brookhaven Protein Data Bank.