BDV matrix protein: from mutation to function
P. Kolenko1, P. Dautel1, R. Novotny2, A.
Martin2, C. Parthier1, M. Schwemmle2, M.T.
Stubbs1
1Institut für Biochemie und Biotechnologie,
Martin-Luther Universität, Kurt-Mothes-Straße 3,
06 120 Halle
(Saale), Germany
2Institut für Virologie, Universität
Freiburg, Hermann-Herder-Straße 11, 79 104 Freiburg, Germany
petr.kolenko@biochemtech.uni-halle.de
Borna disease virus (BDV) is a neurotropic
virus that typically infects horses, sheep and other farm animals [1]. Recent
studies have also shown that genomic BDV-like elements were inserted into the
mammalian genome, including humans [2]. Besides Ebola, Marburg and Vesicular stomatitis virus, BDV belongs to the order of Mononegavirales [3]. One of the six proteins encoded by the
BDV genome (negative stranded non-segmented RNA) is the matrix (M) protein [4].
BDVM occurs dominantly in form of a homo-tetramer with a total molecular weight
of 65 kDa. Monomers of BDVM fold into an L-shaped
β-sandwich consisting of 6 antiparallel strands
that are surrounded by several α-helices (see Fig. 1). BDVM binds
short fragments of RNA and is able to protect them from degradation. Despite a
wide range of studies, the role of BDVM in life cycle of the virus is not fully
understood, yet. We have designed several mutant variants of
the BDVM protein, and analyzed them using analytical size-exclusion
chromatography, analytical ultracentrifugation, RNA-PAGE, spectrometric
methods, and X-ray crystallography. Finally, we performed pairwise comparison of the mutant variants with the wild
type BDVM. We have observed novel oligomeric states,
and also secondary RNA binding sites. According to our experiments, both
aspects play a role in growth of the virus.
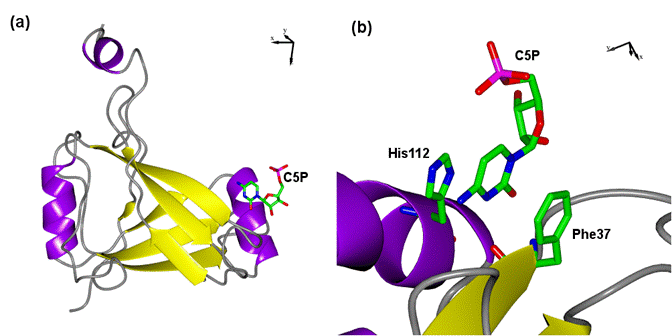
Figure 1. Overall structure of BDVM (a), and its nucleotide binding site (b). BDVM is represented by secondary structure elements, cytidine-5`-monophosphate and interacting residues His112, and Phe37 are represented by sticks. The figure was generated using CCP4MG [5]
1. P.
Staeheli, C. Sauder, J. Hausmann, F. Ehrensperger, M. Schwemmle, J. Gen. Virol.,
81, (2000), 2123-2135.
2. C. Feschotte, Nature, 463,
(2010), 39-40.
3. C.R. Pringle, Archives of Virology, 117, (1991), 137-140.
4. P. Neumann, D. Lieber, S. Meyer,
P. Dautel, A. Kerth, I. Kraus, W. Garten, M.T. Stubbs, PNAS, 106,
(2009), 3710-3715.
5. E. Potterton, S. McNicholas, E. Krissinel, K. Cowtan, M. Noble, Acta Cryst., D58,
(2002), 1955-1957.
Acknowledgements.
This
work was supported by the Deutsche Forschungsgemeinschaft
Graduiertenkollegs 1026 „Conformational transitions
in macromolecular interactions“. The authors wish to acknowledge the use of beamline BL14.1 of BESSY II at the Helmholtz-Zentrum
Berlin and the assistance during data collection.