Solving phase
problem using a Se-Met derivative of the flavoenzyme NAD(P)H:acceptor
oxidoreductase (FerB)
T. Klumpler1,
J. Marek1, V. Sedláček2 , I. Kučera2
1">Laboratory
of Molecular Plant Physiology, Department of Functional Genomics and
Proteomics, Institute of Experimental Biology, Faculty of Science, Masaryk
University, Kamenice 5/A2, CZ 625 00 BRNO, <cny>Czech
Republic</cny></aff>
2Department
of Biochemistry, Faculty of Science, Masaryk University, Kamenice
5/A5, CZ 625 00 BRNO, <cny>Czech Republic</cny></aff></aug>.
klumpler@sci.muni.cz
Keywords: ferric reductase B, FerB, phasing,
MRSAD, MAD
Abstract
The flavin adenine
dinucleotide-dependent enzyme FerB from Paracoccus
denitrificans reduces a range of substrates, including chromate, ferric
complexes, benzoquinones and naphtoquinones. The reduced form of nicotinamide
adenine dinucleotide serves as a source of electrons. Recombinant unmodified
and selenometionine-substituted (Se-Met) FerB derivatives were crystallized,
diffraction data for both forms were collected and the phase problem for Se-Met FerB
dimer was solved by three-wavelength multiple
anomalous dispersion, followed by the combination of molecular replacement and
single-wavelength anomalous diffraction phasing. A molecular-replacement solution
of unmodified FerB tetramer was obtained using Se-Met
structure as a search model.
Introduction
The easiest way to crystal structure determination is
the molecular replacement (MR). Only one native dataset, the coordinates of
homologous structure and software that find the right number, orientation and
translation of initial search model are needed. Two-third of more than 60 000
protein structures deposited in January 2010 in the Protein Data Bank [1] has
been determined using MR techniques. Easy-to-use MR software is available and
over 95% of deposited X-ray structures solved by MR
has been determined using MR protocols of CNS [2], or one of specialized MR
packages AMoRe [3], MOLREP [4] and PHASER [5]. More
recently, software for automatic choice of phasing models from databases has
been released, such as MrBUMP [6] and BALBES [7]. The
solution of phase problem by MR could be very fast in principle, but often
first model requires multiple rounds of manual refinement.
The second most important phasing techniques are based
on intensity differences arising from the presence of heavy atoms implemented
in single or multiple isomorphous replacement with or
without anomalous scattering (SIR/MIR or SIRAS/MIRAS) or in single or multiple
wavelength anomalous diffraction methods (SAD or MAD, [8]). MIR methods were
used to determine the first X-ray structures of macromolecules and still have
potential to determine unknown structure directly from experimental data. Heavy
atoms are introduced to the protein crystal by soaking the crystal in the ionic
solution of heavy atom. On the contrary, SAD and MAD methods incorporate heavy
(typically selenium) atoms into protein crystals using protein molecules
containing selenomethionine instead of methionine [9]. Se-Met methods becomes more and more
popular recently, because they eliminate problems associated with heavy-metal
screening, the lack of isomorphism between native and heavy-atoms structures
and mainly the only one single crystal is needed to perform complete diffraction
experiment. Heavy-atom (or selenium) substructure could be determined using the
Patterson or direct-methods programs such as SnB
[10], SHELXD [11], CNS or SOLVE [12]. Determination of heavy-atom substructure
via SAD can be initialized using preliminary positions of heavy-atom from an MR
solution. This combined technique is called molecular replacement with
single-wavelength anomalous diffraction (MRSAD) [13].
A broad scale of available programs indicates that
crystallographer chooses the most appropriate individual programs for the
specific sub-tasks executed during effective pass through the complete process
of the protein crystal structure determination. At least partial automation of
this multi-step decision process has become recent initiative. Different
automated pipeline have been built up, e.g. ACrS
[14], Auto-Rickshaw [15], autoSHARP [16], CRANK [17],
ELVES [18], HKL-3000 [19], PHENIX [20], SGXPro [21].
The flavin-dependent enzyme FerB from Paracoccus denitrificans reduces a broad range of compounds,
including ferric complexes, chromate and quinones, at
the expense of the reduced nicotinamide adenine dinucleotide cofactors, NADH or NADPH [22, 23]. Enzymes
utilizing flavin cofactors, (flavin
mononucleotide, FMN, or flavin adenine dinucleotide, FAD), are unique in their ability to catalyze
a wide variety of mechanistically different reactions, such as dehydrogenation,
oxygen activation, halogenation, non-redox conversions, light sensing and emission, and DNA
repair [24]. The function of all of these enzymes in cell metabolism has not
yet been fully elucidated. Finding the molecular basis of the catalysis by FerB would be greatly aided by knowledge of the
three-dimensional structure of the enzyme. Here we report the solution of the
phase problem of the Se-Met derivative of flavin
dependent enzyme FerB from Paracoccus denitrificans using the advanced 3W-MAD
and MRSAD protocols of Auto-Rickshaw: the EMBL-Hamburg automated crystal
structure determination platform after MR solution of the native protein FerB had failed.
Materials and methods
1. Production,
purification and crystallization of Se-Met FerB
Se-Met FerB was prepared
using the methionine biosynthesis inhibition method
[25]. Purification in a single chromatography step using a HisPrep
FF 16/10 column (GE Healthcare) [26] resulted in almost homogenous protein
preparations (> 95% homogeneity). Purity and monodisperzity
of a sample were controlled by SDS-PAGE electrophoresis and dynamic light
scattering. For protein crystallization micro-seeding technique was exploited
and crystals of native and Se-Met FerB were obtained
(Fig. 1.). For details see [27].
2.
MALDI-TOF mass spectrometry
Native and Se-Met FerB were
analyzed by matrix-assisted laser desorption/ionization time-of-flight
(MALDI-TOF) mass spectrometry on a ULTRAFLEX III mass spectrometer (Bruker Daltonics, Germany).
Samples were co-crystallized with 2,5-dihydroxybenzoic
acid and analyzed in linear mode using an accelerating voltage of 25 kV. The
instrument was calibrated with [MH]+ and
[MH]2+ peaks using a mixture of peptide standards (Bruker Daltonics). Peaks at
molecular masses of 21289 and 21616 detected for intact and Se-Met FerB correspond well to the predicted mass difference of
328 Da (seven methionine
residues per chain).
3. Auto-Ricksaw structure determination
Se-Met diffraction data were collected at tunable beamline X12 of the DORIS-III storage ring at EMBL/DESY
(Hamburg, Germany), processed and merged using the XDS system [28] (for details
see [27]). The structure of Se-Met FerB was solved
using the advanced 3W-MAD and MRSAD protocols of Auto-Rickshaw: the
EMBL-Hamburg automated crystal structure determination platform. The output
diffraction data from XDS were converted for use in Auto-Rickshaw using
programs of the CCP4 suite (CCP4,1994), and FA values were calculated
using the program SHELXC [11]. Based on an initial analysis of the data, the
maximum resolution for FerB substructure
determination and initial phase calculation was set to 1.8 Ĺ. 14 selenium atoms
were found using the program SHELXD. The correct hand for the substructure was
determined using the programs ABS [29] and SHELXE [11] and initial phases were
calculated after density modification using the program SHELXE. The initial phases
were improved by density modification and phase extension using the program DM
[30]. Then the twofold non-crystallographic symmetry (NCS) operator was found
and then density modification with solvent flattening and NCS averaging was
applied, both using the program RESOLVE [31]. Resulting phases from RESOLVE
were used as input for model building mode of program ARP/wARP.
The model building and refinement protocol implemented
into MRSAD pipeline of Auto-Rickshaw started with rigid-body refinement of individual
protein chains at 4 Ĺ resolution followed by positional, B-factor and once more
positional refinement at 3.0 Ĺ using CNS. The CNS result was used for
refinement and phase extension to 1.75 Ĺ resolution using REFMAC5 [32]. In the
next step of the protocol, quality of electron density map was improved using
density modification and NCS-averaging by RESOLVE (Fig. 2.). New, more complete
model of FerB was prepared consequently building of polyalanine model by beta version of SHELXE, side-chain
docking with RESOLVE, REFMAC5 refinement, and finally by run of ARP/wARP [33, 34] in the model building regime.
Results
We had tried to solve FerB
structure by the molecular replacement (MR) methods using an NAD(P)H
dependent FMN reductase flavoprotein
from Pseudomonas aeruginosa PA01 (PDB code1RTT)
[35] identified by a FASTA search [36]
as a model. Unfortunately, all MR trials were unsuccessful. We therefore
collected data with selenomethionine derivative of FerB. An excellent data quality and their maximum resolution
allow us to try to solve phase problem of FerB using
the advanced 3W-MAD protocol of Auto-Rickshaw: the EMBL-Hamburg automated
crystal structure determination platform. The twofold NCS operator connecting
expected two FerB monomers close to x, -y, ?-z was
found by RESOLVE. The model building mode of program ARP/wARP
employed at the end of the advanced 3W-MAD protocol of Auto-Rickshaw was able
find 309 (from expected 364) residues divided to 11 chains, 100% of them have
been docked in FerB sequence, and this intermediate
model was completed by the MRSAD pipeline of Auto-Rickshaw to almost complete
model of Se-Met FerB homodimer
containing 349 residues divided into 9 chains (340 of them correctly docked)
with final R/Rfree=
0.2144/0.2682.
The structure of native FerB
tetramer was successfully solved by molecular replacement technique implemented
in PHASER (the final translation function Z-score TFZ= 66.6) with manually
unrefined structure of one of Se-Met FerB monomers as
the search model. Refinement of both FerB structures
is now in progress.
Figures
Figure
1. Rod-like crystals of Se-Met FerB.
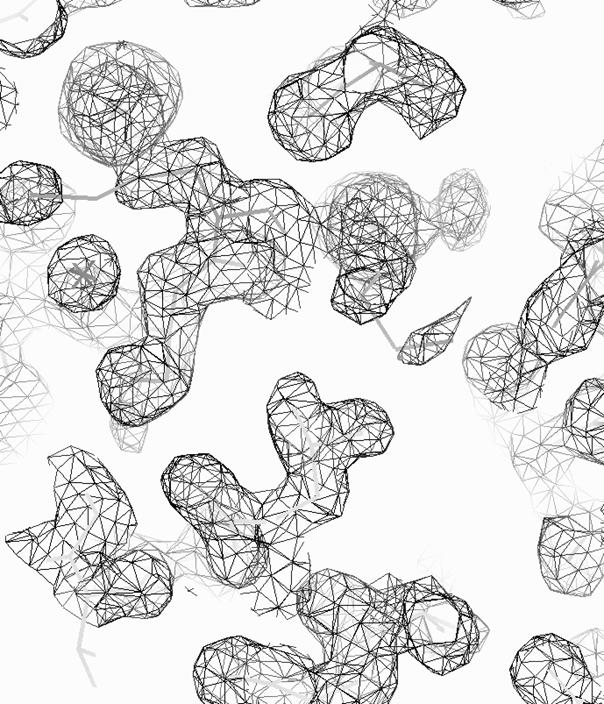
Figure 2. 2FO-FC map of Se-Met FerB contoured at 1σ.
Acknowledgement:
We wish thank to the EMBL/DESY
Hamburg for providing us with synchrotron facilities and D. Tucker for his
assistance with data collection on beamlines X12 and
X13 of the DORIS-III storage ring at DESY Hamburg. The authors are grateful to Ondrej Šedo for measuring the
MALDI-TOF MS spectra and the Meta Center for computer time. This research was
supported by grants from the Czech Science Foundation (grant Nos. 204/08/H054
and 525/07/1069 and P503/10/217) and the Ministry of Education, Youth and
Sports (grant Nos. MSM0021622413, MSM0021622415 and LC06034).
References:
1
H.M. Berman, J.
Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E.
Bourne, Nucleic Acids Res., 28 (2000), 235.
2
A.T. Brünger, P.D. Adams, G.M. Clore, W.L. DeLano, P. Gros, R.W.
Grosse-Kunstleve, J.S. Jiang, J. Kuszewski, M. Nilges, N.S. Pannu, R.J. Read,
L.M. Rice, T. Simonson, G.L. Warren, Acta
Cryst., D54, (1998), 905.
3
J. Navaza, Acta Cryst., A50,
(1994), 157.
4
A. Vagin, A. Teplyakov, J. Appl. Cryst., 30, (1997), 1022.
5
A.J. McCoy, R.W. Grosse-Kunstleve, P.D.
Adams, M.D. Winn, L.C. Storoni, R.J. Read, J.
Appl. Cryst., 40, (2007), 658.
6
R.M. Keegan, M.D. Winn, Acta Cryst., D63, (2007), 447.
7
F. Long, A.A. Vagin, P. Young, G.N.
Murshudov, Acta Cryst., D64, (2008), 125.
8
W.A. Hendrickson, Science, 254, (1991),
51.
9
W.A. Hendrickson, J.R. Horton, D.M.
LeMaster, EMBO J., 9, (1990), 1665.
10
R. Miller, N. Shah, M.L. Green, W.
Furey, C.M. Weeks, J. Appl. Cryst., 40, (2007), 938.
11
G.M. Sheldrick, Acta Cryst., A64, (2008), 112.
12
T.C. Terwilliger, J.
Berendzen, Acta Cryst., D55, (1999), 849.
13
J.P. Schuermann, J.J.
Tanner, Acta Cryst., D59, (2003), 1731.
14
J.S.
Brunzelle, P. Shafaee, X. Yang, S. Weigand, Z. Ren, W.F. Anderson, Acta Cryst., D59, (2003), 1138.
15
S. Panjikar, V. Parthasarathy,
V.S. Lamzin, M.S. Weiss, P.A. Tucker, Acta
Cryst., D65, (2009), 1089.
16
C. Vonrhein, E. Blanc, P. Roversi, G.
Bricogne, Methods Mol. Biol., 364, (2006), 215.
17
S.R. Ness, R.A. de Graaff, J.P.
Abrahams, N.S. Pannu, Structure, 12 (2004), 1753.
18
J. Holton, T. Alber, Proc. Natl Acad. Sci. USA, 101, (2004), 1537.
19
W. Minor, M. Cymborowski, Z.
Otwinowski, M. Chruszcz, Acta Cryst.,
D62, (2006), 859.
20
P.D. Adams, R.W.
Grosse-Kunstleve,L.W. Hung, T.R. Ioerger, A.J. McCoy, N.W. Moriarty, R.J. Read,
J.C. Sacchettini, N.K. Sauter, & T.C. Terwilliger, Acta Cryst., D58,
(2002), 1948.
21
Z.Q. Fu, J. Rose, B.C. Wang, Acta Cryst., D61, (2005), 951.
22
J. Mazoch, R. Tesařík, V. Sedláček, I.
Kučera, J. Turánek, Eur. J. Biochem.,
271, (2004), 553.
23
V. Sedláček, R.J.M. van Spanning, I.
Kučera, Arch. Biochem. Biophys., 483, (2009), 29.
24
A. Mattevi, Trends Biochem. Sci., 31,
(2006), 276.
25
G.D. Van Duyne, R.
Standaert, P.A. Karplus, S.L. Schreiber, J. Clardy, J. Mol. Biol.,
229, (1993), 105.
26
R. Tesařík,
V. Sedláček, J. Plocková,
M. Wimmerová, J. Turánek,
I. Kučera, Protein
Expres. Purif., 68,(2009),
233.
27
T. Klumpler, V. Sedláček, J. Marek, M.
Wimmerová, I. Kučera, Acta Cryst., F66, (2010), (in press).
28
W. Kabsch, J. Appl. Cryst., 26, (1993), 795.
29
Q. Hao, J. Appl. Cryst., 37, (2004), 498.
30
K. Cowtan, Joint CCP4 and ESF-EACBM Newsletter on protein crystallography, 31, (1994), 34.
31
T.C. Terwilliger, Acta Cryst., D59,
(2003), 38.
32
G.N. Murshudov, A.A. Vagin, E.J.
Dodson, Acta Cryst., D53, (1997), 240.
33
A. Perrakis, R.J. Morris, V.S. Lamzin, Nature Struct. Biol., 6, (1999), 458.
34
S.X. Cohen, M. Ben Jelloul, F. Long, A.
Vagin, P. Knipscheer, J. Lebbink, T.K. Sixma, V.S. Lamzin, G. N. Murshudov, A.
Perrakis, Acta Cryst., D64, (2008), 49.
35
R. Agarwal, J.B. Bonanno, S.K. Burley,
S. Swaminathan, Acta Cryst., D62, (2006), 383.
36
W.R. Pearson, D.J. Lipman, Proc. Natl. Acad. Sci. USA, 85, (1998), 2444.