How is a-rna treated by
different force fields and salt conditions?
Ivana
Beššeová,1,2 Michal Otyepka,1,3 Kamila Réblová1 and Jiří Šponer1
1Institute of Biophysics, Academy of Sciences of the Czech Republic, Královopolská 135
612 65, Brno, Czech Republic.
2Gilead Sciences & IOCB
Research Center, Institute of Organic Chemistry and Biochemistry, Academy of
Sciences of the Czech Republic, Flemingovo náměstí. 2, 166 10, Prague 6,
Czech Republic
3Department of Physical
Chemistry and Centre for Biomolecular and Complex Molecular Systems, Palacký
University, tr. Svobody 26, 771 46 Olomouc, Czech Republic
i.besse@mail.muni.cz
An
extensive molecular dynamics study
The
A-RNA duplexes were more compact when using the Parmbsc0 force field compared
to the Parm99. In addition, the Parmbsc0 temporarily reduced α/γ t/t flips. Nevertheless,
since the α/γ
t/t sub-state occurs
to a certain extent in experimental A-RNA structures, we consider both force
fields as viable. The effects of the Parmbsc0 force field included visible reduction
of the major groove width, increase of the base pair roll, larger helical
inclination and small increases of twist. The Parmbsc0 shifted the simulated duplexes
more deeply into the A-form. [3]
A
narrowing of the deep major groove was observed in excess salt simulations, again
accompanied by larger roll, inclination and twist. [3]
The
differences between Parm99/lower-salt and Parmbsc0/higher-salt Parmbsc0 conditions
were small; nevertheless their cumulation induced visible stabilization of the
A-RNA helices. In addition, the effects of the force field and salt conditions were
sequence-dependent. Thus, the compactness of A-RNA is sensitive to the sequence
and the salt strength which may, for example, modulate the end-to-end distance
of the A-RNA helix.
1.
J. M. Wang, P. Cieplak and P. A. Kollman, J. Comput.
Chem., 21
3.
I. Besseova, M. Otyepka, K. Reblova and J. Sponer, Phys.
Chem. Chem. Phys., 11
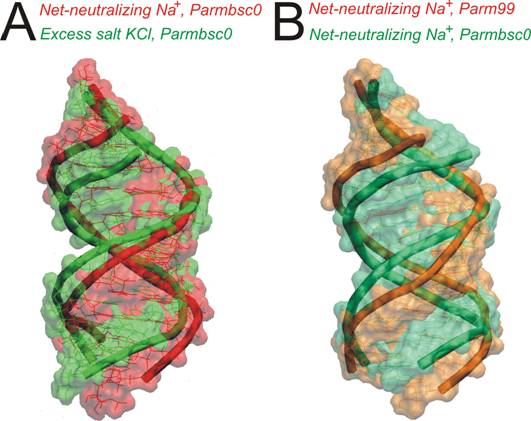
Figure
1. A-RNA simulations: ionic conditions influence the major groove width
and compactness of the helix.