Molecular dynamics study: Intrinsic dynamics of RNA three-way junction
I. Beeová1,2, K. Réblová1, J. poner1
1 Institute
of Biophysics, Academy of Sciences of the Czech Republic, Královopolská 135,
612 65 Brno, Czech Republic
2 Gilead Sciences&IOCB Research Center, Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic, Flemingovo náměstí. 2, 166 10, Prague 6, Czech Republic
i.besse@mail.muni.cz
Ribosome is a large biomolecular machine
which resembles a brick-box composed of molecular building blocks - RNA motifs having different shapes, flexibilities, capabilities to
interact with ribosomal surrounding elements, e.g K-turns possessing elbow
dynamics [1], RNA helix showing anisotropic bending [2] or segment of specific
rigid shape [3]. Molecular dynamics (MD) simulation is a suitable method for
characterizing properties and topology of individual RNA segments.
This research is focused on dynamics of one of the important ribosomal
building block - RNA three-way junction (3WJ)
family C. [4] The structure is composed of three helices P1, P2 and P3
diverging from one point, while P1 and P2 helices are coaxially stacked. [4]
There are also tertiary interactions between stems P1 and P3, which are
characteristic especially for the family C. [4] The study involves three
junctions -
This extensive molecular dynamics study
including simulations of total length more than 0.6 μs showed two dominant
structural motions which are very similar for all three 3WJs. The first
dynamics includes hinge-like fluctuations between the coaxially stacked stems
P1/P3 (forming a compact RNA part of the structure) and P2, classified as a
hinge-like motion (see Figure 1 B-C). The second one is internal dynamics of
stems P1 and P3 called as a breathing-like motion (see Figure
All three studied junctions are associated with extended regions of negative electrostatic potential which are in many cases major binders of monovalent cations with 100% occupancy and very slow exchange of ions.
To sum up, 3WJs belong to RNA building blocks including a characteristic intrinsic dynamics of hinge- and breathing- like motion.
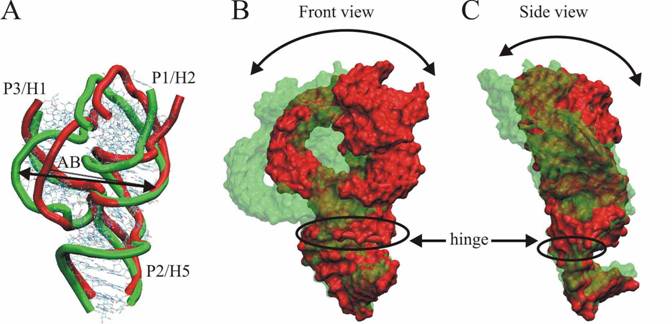
Figure
1 Overlay of 5S 3WJ structures in its extremal substates of two characteristic intrinsic dynamics
(A) breathing- like motion, (B, C) hinge- like motion.
References
2. K. Reblova, F. Lankas, F. Razga, M.V. Krasovska, J. Koca and J.
Sponer, Biopolymers, 82 (2006) 504-520.
3. N. Spackova and J. Sponer, Nucleic Acids Research, 34 (2006)
697-708.