Theoretical NMR Study of Watson-Crick/Sugar Edge RNA Base Pair Family
Z. Vokáčová1, J. Šponer2, V. Sychrovský1
1Institute of Organic Chemistry and
Biochemistry, Academy of Sciences of the Czech Republic, Flemingovo square 2,
166 10 Prague 6, Czech Republic
2 Institute of
Biophysics, Academy of Sciences of the Czech Republic, Královopolská 135, 612
65 Brno, Czech Republic
Ribonucleic acid, one type of nucleic acids, has large variety of forms and also shows an astonishing variability of base-pairing [1,2].
Classical Watson-Crick type of base pairing found in DNA represents only 50% of base pairing in RNA statistically[1,2]. There are many non-canonical base pairs in RNA that have no counterpart in DNA. A role of the non-WC base pairs in RNA is fundamental since they participate in folding and stabilization of RNA tertiary structure. Detection of binding motives in rather complicated and highly variable RNA macromolecules can help in better understanding of chemical processes of nucleic acids.
Each nucleoside possesses three edges [1] shown in Figure 1: WC edge, Hoogsteen edge (for purines) or CH edge (for pyrimidines), and Sugar edge (SE). A given edge of one nucleoside can in principle interact with one of the three edges of a second nucleoside. This interaction can be either cis or trans with respect to sugar moiety. According to Leontis and Westhof [1], all possible combinations lead to twelve families of distinct geometry patterns.
Quantum chemistry study of nuclear magnetic resonance parameters of all members of cis- and trans- Watson-Crick/Sugar edge base pair family was provided by mean of calculation of indirect spin-spin coupling constants with the CP-DFT method. RNA base pair interact via hydrogen bonds. Inter and intra indirect spin-spin couplings of local binding motif are calculated and are compared to experimental data and obtained structural dependences of spin-spin couplings are described. Forty-two complexes are studied and effects of local structural modification, water mediation and solvent effect are calculated. Each H-bond pattern has its own set of representative J-coupling which can be used for the identification of particular RNA bonding pattern via NMR spectroscopy.
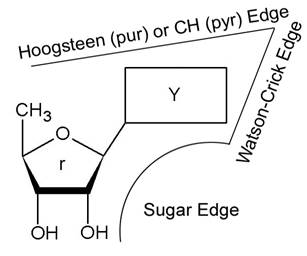
Figure 1. Classification of
the interaction edges for RNA nucleoside by Leontis [1]
References
1. N. B. Leontis and E. Westhof, RNA, 7 (2001) 499-512.
2.
N. B. Leontis, J. Stombaugh and
E. Westhof, NAR, Vol. 30 No.16
(2002) 3497-3531