Crystallization
study of three mutant haloalkane dehalogenases derived
from dehalogenase DhaA of Rhodococcus
rhodochrous NCIMB 13064
Alena Stsiapanavaa, Tana Koudelakovac,
Lucie Grodeckac, Jiri Damborskyc, and Ivana Kuta Smatanovaa,b
aInstitute
of Physical Biology University of South Bohemia Ceske Budejovice, Zamek 136,
373 33 Nove Hrady, Czech Republic
bInstitute
of Systems Biology and Ecology Academy of Science of the Czech Republic, Zamek
136, 373 33 Nove Hrady, Czech Republic
cLoschmidt
Laboratories, Faculty of Science, Masaryk University, Kamenice 5/A4, 62500
Brno, Czech Republic
Haloalkane dehalogenases (EC 3.8.1.5) are
enzymes that belong to the α/β-hydrolase fold family. These microbial
enzymes catalyze hydrolytic conversion of halogenated hydrocarbons to
corresponding alcohols [1]. Dehalogenation is a key step in aerobic
mineralization pathways of many halogenated compounds that occur as
environmental pollutants [2]. Haloalkane dehalogenases are potentially
important biocatalysts with both industrial and bioremediation applications
that could be used for industrial biocatalysis or as active compounds of
biosensors, respectively [3, 4].
Wild-type DhaA was isolated from bacterium Rhodococcus
rhodochrous NCIMB 13064
[5]. Derived mutant enzymes DhaA04, DhaA14 and DhaA15 were constructed to
reveal importance of product transporting pathways (tunnels) in DhaA for its
enzymatic activity. Our project is aimed to produce crystals of haloalkane
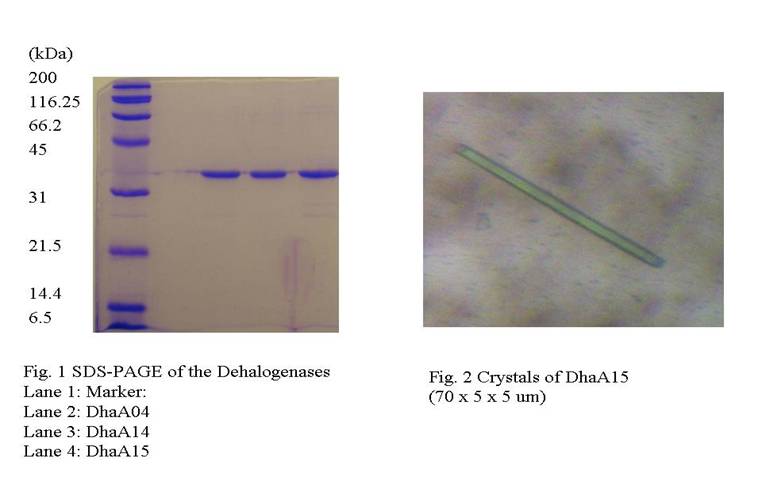
dehalogenases DhaA04, DhaA14 and DhaA15 purified mutants (Fig. 1) in efficient
quality for diffraction experiments and finally compare results with known
structure of wild-type DhaA [3].
Standard vapor diffusion technique has been
used for searching and optimization of crystallization conditions.
Crystallization experiments have been performed in Hampton Research Linbro and
Cryschem plates (Hampton Research, CA, USA) as well as in Emerald BioStructures CombiClover Crystallization Plate
(EBS plate, Emerald BioStructures, WA,
USA) using commercial crystallization kits as Crystal
Screen Lite and Crystal Screen of Hampton Research, and Clear Strategy Screen 1
of Molecular Dimensions Limited (MDL, Suffolk, UK). The first microcrystals of
DhaA15 were obtained from PCT, reagent B2 of Hampton Research (Fig. 2).
Crystallization experiments with all
enzymes are in the progress.
Acknowledgements:
This work is supported by the
Ministry of Education of the Czech Republic (MSM6007665808 and LC06010) and by
the Academy of Sciences of the Czech Republic (AVOZ60870520).
References:
1. Dick B Janssen: Evolving Haloalkane
Dehalogenases. Current Opinion in Chemical Biology, 8, 2004, 150‑159
2. Dick B Janssen, Inez J. T. Dinkla, Gerrit
J. Poelarends and Peter Terpstra: Bacterial degradation of xenobiotic
compounds: evolution and distribution of novel enzyme activities. Environmental Microbiology, 7, 2005, 1868‑1882.
3. Janet Newman, Thomas S. Peat, Ruth Richard, Lynn Kan, Paul E. Swanson, Joseph A.
Affholter, Ian H. Holmes, John F. Schindler, Clifford J. Unkefer and Thomas C.
Terwilliger: Haloalkane Dehalogenases: Structure of a Rhodococcus Enzyme. Biochemistry,
38, 1999, 16105‑16114
4. Zbynek Prokop,
Marta Monincova, Radka Chaloupkova, Martin Klvana, Yuji Nagata, Dick B. Janssen
and Jiri Damborsky: Catalytic Mechanism of the Haloalkane Dehalogenases Lin B
from Sphingomonas paucimobilis UT26*.
The Journal of Biological Chemistry, 46,
2003, 45094‑45100
5. Anna N. Kulakova, Michael J. Larkin and Leonid A. Kulakov: The plasmid-located haloalkane dehalogenase gene from Rhodococcus
rhodochrous NCIMB 13064. Microbiology, 143, 1997, 109–115
![]()