Interaction of Forkhead Transcription Factor FoxO4 with DNA
Tomáš Obšil1,2
1 Department of Physical and Macromolecular Chemistry, Faculty of Science, Charles University; 12843 Prague, Czech Republic
2 Institute of Physiology, Academy of Sciences of the Czech Republic; 14220 Prague, Czech Republic
FoxO4 belongs to the “O” subset of forkhead transcription factors, which participate in various cellular processes. The forkhead DNA binding domain (DBD) consists of three-helix bundle resting on a small antiparallel β-sheet from which two extended loops protrude and create two wing-like structures (Fig. 1). The main DNA recognition site is a-helix H3 that makes contacts with the major groove of DNA. Other regions of forkhead domain that can make important interactions with DNA are both wings or N-terminal extension upstream of helix H1 [1,2].
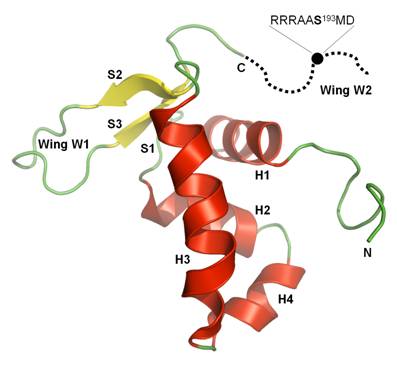
Fig. 1: The ribbon representation of solution structure of FoxO4–DBD sequence Ser92-Gly181 [3]. The missing part of wing W2 is schematically shown as dotted line. Black circle represents approximate location of PKB phosphorylation site Ser193.
Within the large family of Fox transcription factors, the proteins FoxO1 (FKHR), FoxO3a (FKHR-L1), FoxO4 (AFX) and FoxO6 constitute the “O” subfamily. Members of this subfamily play an important role in cellular proliferation, survival, and in mediating effects of insulin and growth factors on metabolism [4]. All FoxO proteins function under the control of the phosphoinositide-3-kinase–protein kinase B (PI3K–PKB) pathway. Phosphorylation by PKB creates two binding sites for the 14-3-3 protein and induces phosphorylation of additional sites by casein kinase 1 and dual-specificity tyrosine(Y) regulated kinase 1A. PKB-mediated phosphorylation induces binding of the 14-3-3 protein and the resulting complex is then translocated to the cytosol where the bound 14-3-3 protein prevents re-entry of FoxO into the nucleus likely by masking its nuclear localization sequence [5-7].
The DNA binding potential of FoxO–DBD is controlled by multiple mechanisms. The most important factors seem to be the PKB-induced phosphorylation and the binding of the 14-3-3 protein [6-9]. PKB phosphorylation site and the 14-3-3 protein binding motif are located in the basic region of wing W2 at the C-terminus of FoxO–DBD (Fig. 1). Structures of HNF-3g (FoxA3) and Genesis (FoxD3) complexes revealed that cluster of basic residues in the wing W2 is involved in DNA binding [1,10]. Moreover, the removal of wing W2 abolishes DNA binding of forkhead transcription factor HNF-3α. Sequence alignment between HNF-3g, Genesis and FoxO sequences suggests that analogues basic residues forming the second FoxO PKB phosphorylation site and the 14-3-3 binding motif might participate in DNA binding as well. This similarity could explain the inhibitory effect of both the phosphorylation and the 14-3-3 protein on DNA binding activity of FoxO proteins. However, the regulation of DNA binding among various FoxO proteins seems to differ significantly. It has been shown that phosphorylation of the second PKB/14-3-3 binding motif in the wing W2 suppresses DNA binding of FoxO1 and FoxO6 factors [9]. On the other hand, the PKB-induced phosphorylation of both DAF-16 (Caenorhabditis elegans FoxO homologue) and FoxO4 (fragment 11-213) does not by itself affect their binding to the target DNA [6,8]. Binding of the 14-3-3 protein to phosphorylated FoxO4 and DAF-16 has been shown to be necessary for complete inhibition of their binding to the DNA.
To better understand these differences among FoxO–DBD, we investigated the role of N-terminal loop (portion located upstream of first helix H1) and C-terminal region (loop known as wing W2) of forkhead domain of transcription factor FoxO4 in DNA binding. While the deletion of either portion partly reduces the FoxO4–DBD binding to the DNA, the simultaneous deletion of both regions inhibits DNA binding significantly. Förster resonance energy transfer measurements and molecular dynamics simulations suggest that both studied N- and C-terminal regions of FoxO4–DBD directly interact with DNA. In the presence of N-terminal loop the PKB-induced phosphorylation of wing W2 by itself has negligible effect on DNA binding. On the other hand, in the absence of this loop the phosphorylation of wing W2 significantly inhibits the FoxO4–DBD binding to the DNA. The binding of the 14-3-3 protein efficiently reduces DNA binding potential of phosphorylated FoxO4–DBD regardless of the presence of N-terminal loop. Our results show that both N- and C-terminal regions of forkhead domain are important for the stability of FoxO4–DBD/DNA complex [11].
References
[1] K. L. Clark, E. D. Halay,
[2] N. Zheng,E. Fraenkel, C.O. Pabo, N.P. Pavletich, Genes Dev., 13 (1999) 666-674.
[3] J. Weigelt, I. Climent, K. Dahlman-Wright, M. Wikstrom, Biochemistry, 40 (2001) 5861-5869.
[4] B.M. Burgering, G.J. Kops, Trends Biochem. Sci., 27 (2002) 352-360.
[5] A. Brunet, A. Bonni, M.J. Zigmond, M.Z. Lin, P. Juo, L.S. Hu, M.J. Anderson, K.C. Arden, J. Blenis, M.E. Greenberg, Cell, 96 (1999) 857-868.
[6] T. Obsil, R. Ghirlando, D.E. Anderson, A.B. Hickman, F. Dyda, Biochemistry, 42 (2003) 15264-15272.
[7] V. Obsilova, J. Vecer, P. Herman, A. Pabianova,
[8] C.M. Cahill, G. Tzivion, N. Nasrin, S. Ogg, J. Dore, G. Ruvkun, M. Alexander-Bridges, J. Biol. Chem., 276 (2001) 13402-13410.
[9] X. Zhang, L. Gan, H. Pan, S. Guo, X. He, S.T. Olson, A. Mesecar, S. Adam, T.G. Unterman, J. Biol. Chem. 277 (2002) 45276-45284.
[10] C. Jin, I. Marsden, X. Chen, X. Liao, J. Mol. Biol., 289 (1999) 683-690.
[11] E. Boura, J. Silhan, P. Herman, J.
Vecer,
Acknowledgement
This work was funded by Grant 204/06/0565 of the Grant Agency of the Czech Republic, by Grant KJB500110601 of the Grant Agency of the Academy of Sciences of the Czech Republic, by Research Project MSM0021620835 and Centre of Neurosciences LC554 of the Ministry of Education, Youth, and Sports of the Czech Republic, and by Research Project AV0Z50110509 of the Academy of Sciences of the Czech Republic.