Structure and dynamics of the oxygen evolving complex of photosystem II: Role of the N-terminal loop of PsbQ
Rüdiger Ettrich1, Jaroslava Ristvejova1, Vladimír Kopecky jr.2, Žofie Sovova1, Kateřina Hofbauerová3, Juan B. Arellano4
1Laboratory of High Performance Computing, Institute of Systems Biology and Ecology ASCR and Institute of Physical Biology USB, Zamek 136, 37333 Nové Hrady, Czech Republic.email:ettrich@greentech.cz
2Institute of Physics, Faculty of Mathematics and Physics, Charles University, Ke Karlovu 5, 12116 Prague 2, Czech Republic
3Institute of Microbiology, Academy of Sciences of the Czech Republic, Vídeňská 1083, CZ-14220 Prague 4, Czech Republic
4Instituto de Recursos Naturales y Agrobiología (CSIC), Cordel de Marinas 52, 37008 Salamanca, Spain
Infrared and Raman spectroscopy were applied to identify restraints for
the structure determination of the 20 amino acid loop between two beta-sheets
of the N-terminal region of the PsbQ protein of the oxygen evolving complex of
photosystem II from Spinacia oleracea by restraint-based homology
modeling. One of the initial models has shown a stable fold of the loop in a
20 ns molecular dynamics simulation that is in accordance with
spectroscopic data. Cleavage of the first 12 amino acids leads to a permanent
drift in the root means square deviation of the protein backbone and induces
major structural changes.
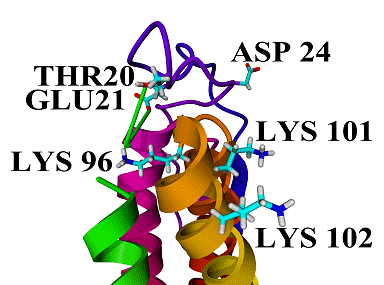
Figure
1: Lys96, one of the four lysyl residues which are probably orientated to the
lumenal facing intrinsic proteins of PSII, lies on the opposite side as are the
other three lysyl residues and the conserved loop residue Asp24, in a distance
from 6–12 Å and 7–13 Å to the loop residues Thr20 and
Glu21, respectively. in the MD simulation
The
probable binding site of PsbQ to the complex could be formed by the lysyl rich
region of the helix bundle and the N-terminal loop region around Asp24 and thus
would contain a large positively charged region and a small negatively one. We
hypothesize that after binding to PSII the loop loses its high flexibility and
bends in the direction of Lys96 with Thr20 and Glu21 interacting with this
residue and so burying it under the accessible surface (Fig. 1). Thus
Lys96 could probably behave as a molecular hook holding Glu21 by a salt bridge.1
Acknowledgements.
Supports
from the Institutional Research Concept of the Academy of Science of the Czech
Republic (No. AVOZ60870520) and from the Ministry of Education of the Czech
Republic (No. LC 06010, No. MSM0021620835, No. MSM6007665808) are gratefully
acknowledged. This work was also funded by the Spanish Ministry of Education
and Science (Project Ref.: BFU2004-04914-C02-02/BMC).
1. Ristvejova, J., Kopecky, V., Sovova, Z., Balsera, M., Arellano, J.B., Green, M., Ettrich, R., Biochem. Biophys. Res. Comm., 2006, 345 (1), 287-291.