DISCOVERY OF STEREOSELECTIVE HALOALKANE DEHALOGENASE: NEW TOOL FOR ASYMMETRIC SYNTHESIS
Z. Prokop1, 2, Y. Sato3,
T. Mozga1, D.B. Janssen4, Y. Nagata3 and J. Damborsky1
1Loschmidt
Laboratories, Masaryk University, Kamenice 5/A4, 625 00 Brno, Czech Republic
2National Centre for Biomolecular Research, Masaryk University,
Kamenice 5/A4, 625 00 Brno, Czech Republic
3Department of Life Sciences, Graduate School of Life Sciences, Tohoku University, 2-1-1 Katahira, Sendai, 980-8577, Japan
4Department of Biochemistry, University of Groningen, Nijenborgh 4, 9747 AG Groningen, The Netherlands
The spectrum of classical synthetic methods is significantly widened and enriched by expansion of biochemical methods. Enzyme-catalyzed reactions have become popular alternatives to classical organic chemistry for its high selectivity and activity under mild reaction conditions and several industrial processes using enzymes as a catalyst are already in use. Haloalkane dehalogenases (EC 3.8.1.5) are one class of such enzymes holding high potential for application in asymmetric biocatalysis. The main products of haloalkane dehalogenase reactions (alcohols) are valuable building blocks in organic synthesis and these enzymes do not need any cofactor or metal ion for their activity [1]. However, there has been no report that a member of this specific enzyme family shows sufficient stereoselectivity for production of optically active compounds. In 2001, Pieters and co-workers [2] have investigated enantioselectivity of haloalkane dehalogenase DhlA from Xanthobacter autotrophicus GJ10 and DhaA from Rhodococcus rhodochrous NCIMB 13064. The magnitude of the chiral recognition was low; a maximum E-value of 9 was reached after a structural optimization of the substrate. In 2004, twenty years after discovery of the first haloalkane dehalogenase, development of enantioselective dehalogenases for use in industrial biocatalysis was defined as one of the major challenges of the field [3].
Hydrolytic dehalogenation of a serie of racemic substrates by using three different enzymes has been performed in this study. The enzymes included in testing were previously studied DhaA, and two additional enzymes LinB from Sphingobium japonicum UT26 [4] and DbjA from Bradyrhizobium japonicum USDA110 [5]. The magnitude of the chiral recognition was low for most of the substrates, however excellent enantioselectivity of all three enzymes was observed in reaction with brominated esters of propionic and butyric acids. This observation demonstrated for the first time that a member of haloalkane dehalogenase family possesses sufficient enantioselectivity for synthesis of optically pure compounds. Furthermore, haloalkane dehalogenase DbjA showed high enantioselectivity also with b-substituted bromoalkanes. This enantioselectivity is interesting and surprising in light of a simple structure of this compound giving a very few possibilities for differential binding and catalysis of its enantiomers. On the other hand, b-substituted esters have an extra group that can form additional hydrogen bond with the active site residues and make their binding more oriented. The resolution of esters could be achieved by three-point-attachment mechanism, which would not however apply to b-substituted alkanes (Figure 1). A hypothesis of two separate mechanisms of haloalkane dehalogenase enantioselectivity towards b-substituted bromoalkanes and b-substituted esters has been proposed based on these observations. The extended substrate mapping together with molecular modeling brought the first view on mechanism of haloalkane dehalogenase enantioselectivity. Understanding of these mechanisms is essential for engineering of new enantioselective biocatalysts.
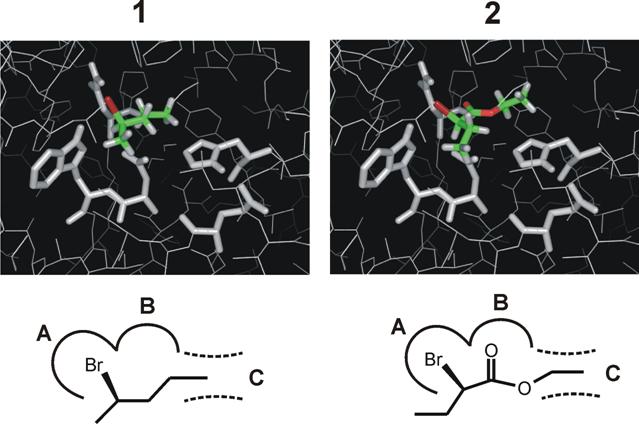
Figure 1 Active-site model of the haloalkane
dehalogenase DbjA. Binding of two types of substrate molecules: b-substituted alkanes (1) and b-substituted esters (2). A,
halide-binding site; B, oxygen-interacting site; C, alkyl-chain binding site.
Catalytic pentad (in white) and substrate molecules docked in Michaelis complex
(in color) are shown in stick.
1. D. B. Janssen, F.Pries & J. R. Van der Ploeg, Annual Review of Microbiology, 48 (1994) 163-191.
2. R. J. Pieters, J. H. L. Spelberg, R. M. Kellogg & D. B. Janssen, Tetrahedron Letters, 42 (2001) 469-471.
3. D. B. Janssen, Current Opinion in Chemical Biology, 8 (2004) 150-159.
4. Y. Nagata, K. Miyauchi, J. Damborsky, K. Manova, A. Ansorgova & M. Takagi, Applied and Environmental Microbiology 63, (1997) 3707-3710.
5. Y. Sato, M. Monincova, R. Chaloupkova, Z. Prokop, Y. Ohtsubo, K. Minamisawa, M. Tsuda, J. Damborsky & Y. Nagata, Applied and Environmental Microbiology, 71 (2005) 4372-4379.