Computer-Assisted Design of Fluorescent Probes for Solvent Relaxation Experiments
1Jeřábek,
P., 1Brezovský, J., 2Hof,
M., 1Damborský, J.
1Loschmidt Laboratories, Masaryk University, Brno, Czech Republic
2 J. Heyrovsky Institute of Physical Chemistry, Academy of Sciences of the Czech Republic, Prague, Czech Republic
The presence and dynamics of solvent molecules may have considerable impact on binding affinities and catalytic rates of enzymes. Solvent relaxation experiments can be used to study solvent dynamics to molecular level. A fluorescent probe with good affinity for the active site of a protein is required for these experiments. Objective of this study was to design probe molecules with appropriate spectral and binding properties.
Dehalogenation reaction proceeds in the three reactions steps – nucleophilic substitution, nucleophilic addition and elimination, during which a substrate is converted to a product. A mutant form of the enzyme with inhibited nucleophilic addition preventing a probe leaving from the active site should be constructed and used for the experiments. This will make possible to study the solvent motion around the probe with fluorescence spectroscopy. Three haloalkane dehalogenase enzymes were selected and genetically modified LinB from bacterium Sphingomonas paucimobilis UT26, DhaA from bacterium Rhodoccoccus sp. NCIMB13064 and DbjA from bacterium Bradyrhizobium japonicum USDA110. Crystal structures of these enzymes were used to construct single-point mutants of a catalytic histidine to phenylalanin.
Molecular docking was then used to design probes with appropriate properties. The main task of this study was to find an appropriate length of reactive linker (Figure 1), which is responsible for location of fluorescent part of the probe inside the enzyme. This part of a probe can be positioned in the active site, in the tunnel joining the active site with protein surface or can stick out from the enzyme to a bulk solvent. According to position of fluorescent probe one can potentially study solvent dynamics in different parts of an enzyme. Probes with different length of reactive linkers were systematically modelled into individual enzymes. The prodan molecule was selected as a fluorescent part of probes (Figure 1). Several fluorescent probes with different position of prodan part inside the enzyme active site were designed. These probes will be syntheised and tested experimentally.
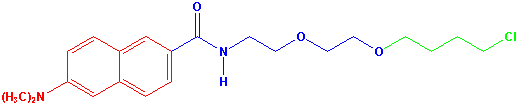
Figure
1. Structural
formula of a fluorescent probe. The fluorescent part of molecule is in red, the
reactive linker is in blue and the substrate part is in green.