The use of GPU based calculations in structural biology
M. Hušák
Department of Solid State, Institute of Chemical Technology Prague,Technická 5, 166 28 Prague 6, Czech Republic
husak@vscht.cz
Keywords: GPU, acceleration, computation, graphic card
There still exist a lot of very computational extensive task in both structural biology and crystallography. Typical examples of problem requesting extreme computational power are QM calculation, molecular dynamic simulations or structure solution from powder data.
The latest development in computer graphic HW gives us an alternative to the classical computational model based on single CPU or clusters of PC.
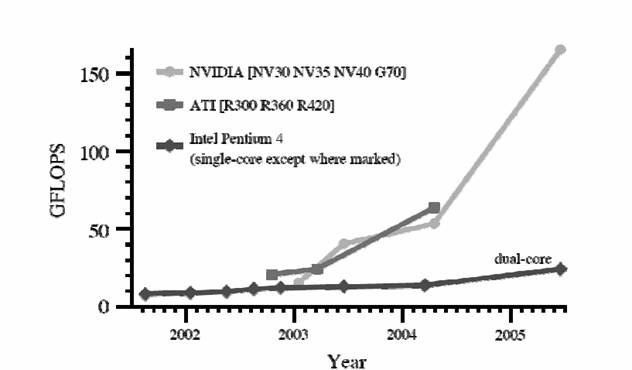
Figure 1. Evolution of the GPU speed in comparison to CPU speed [1].
The Fig.1 demonstrates clearly the nowadays differences between the computational power of graphic cards and single core or dual core standard CPU. The trend is clear – the already existing graphic cards have up to 20 times higher brute force computational power in comparison to existing CPU. The main reason of such a speed difference is computation distribution between multiple GPU processors inside the graphic card and the use of very fast memory elements as well as support for HW implemented vector instructions.
Unfortunately utilization of GPU for normal calculations is non trivial. The algorithm must operate with the multi processor structure of the GPU as well as with several data operation limitations inside the graphic card. In the area of structural biology the GPU based calculations were experimentally used for solving of following problems: Molecular dynamic simulation [2], HMM Viterby probability search in structure databases [3] and NMR-distance smoothing for protein structure determination [4].
In our department we had made only some proof of the concept calculation. We had repeated the NMR-distance smoothing algorithm [3] benchmarks and we had made very simple benchmarks based on several trivial mathematic calculations.
Generally the GPU based calculation is a promising tool for drastic speed up of important computations. To fully utilize it for the structural biology needs it is necessary to modify existing algorithm to versions able to use parallel processing. There still exist several principal limits related to the nature of the graphic HW making GPU non suitable for several key operations (as e.g. large linear equations system solving). The reason is low computation precision in GPU and different data caching model in comparison to CPU. It is expected that such problems will be handled in new graphic cards optimized not only for graphic operations, but also for potential use as computational units.
1. A. Lefohn, I. Buck, J. Owens, T. Purcell, P. McCormick,R. Strzodka. , IEEE Visualization 2005
2. I. Buck, V. Vaidyanathan, V. Rangasayee, V. Pande, E. Darve , ASC Review October 2004
3. D. R. Horn, M. Houston, P. Hanrahan, IEEE Supercomputing 2005
4. P. Micikevicius, in GPU Gems 2, edited by M. Pharr (Addison-Wesley), 2005, Ch. 43
Acknowledgements.
This study was part of research
programme MSM 6046137302 Preparation and research of functional materials and
material technologies using micro- and nanoscopic methods