TRITON: graphic software for modelling protein mutants and calculation reaction pathways
Martin Prokop, Jiří
Damborský, Jaroslav Koča
National Centre for Biomolecular Research, Faculty of
Science, Masaryk University, Kotlářská 2, 611 37 Brno, Czech Republic
One of the objectives of protein engineering is to propose and construct modified enzymes with improved catalytic activity for substrate of interest. The rational engineering of an enzyme requires to know which amino acid residues of the protein are involved in the catalysis and how to modify them to achieve an increased activity.
The program TRITON is a graphical tool for modelling protein mutants and assessment of their activities [1,2]. Protein mutants are modelled based on the wild type structure by homology modelling using the external program MODELLER. Enzymatic reactions taking place in the mutants active site are modelled using the semi-empirical quantum mechanic program MOPAC. Activities of the mutants can be estimated by evaluation the changes in energies of the system and partial atomic charges of the active site residues during the reaction. The program TRITON offers graphical tools for preparation of input data files, for calculation and for the analysis of generated output data. Implementation ensures overall integrity of consecutive steps of the modelling of mutants and calculation of reaction pahways. The program and its methodology were proven by several studies performed on haloalkan dehalogenase enzymes [3-5]. Calculated results showed qualitative agreement with experimental data.
The program TRITON can run under operating system IRIX, Linux and NetBSD. The software is available at http://ncbr.chemi.muni.cz/triton/triton.html
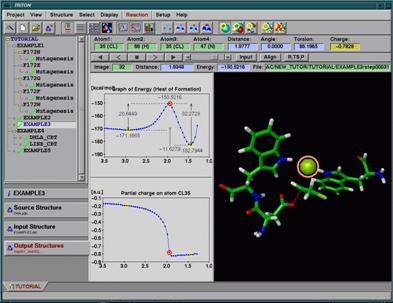
Fig. 1. The main window of the program TRITON. The tree list of the projects and folders for access to the data of specific subprojects are situated on the left. The plot of the reaction coordinate versus the potential energy is situated in the middle-top window and the plot of the reaction coordinate versus partial atomic charge on a selected atom in the middle-bottom window. A 3D model of the structure is in the right window.
1. M. Prokop, J. Damborský, J. Koča, Bioinformatics, 16 (2000) 845-846.
2. J. Damborský, M. Prokop, J. Koča, Trends Biochem. Sci., 26 (2001) 71-73.
3. J. Damborský, M. Boháč, M. Prokop, J. Koča, Prot. Engng., 11 (1998) 901-907.
4. M. Boháč, Y. Nagata, Z. Prokop, M. Prokop, M. Monincová, M. Tsuda, J. Koča, J. Damborský, Biochemistry, 41 (2002) 14272-14280.
5. A. Oakley, M. Klvaňa, M. Otyepka, Y. Nagata, M.C.J. Wilce, J. Damborský, Biochemistry, 43 (2004) 870-878.