Molecular dynamics study: Cationic phosphoramidate a-oligonucleotides targeting single-stranded DNA and RNA efficiently stabilized by butyl amino/guanidium tethers
I. Barvík Jr.
Charles University, Faculty of Mathematics and Physics, Institute of Physics, Ke Karlovu 5, Prague 2, 121 16, Czech Republic
Much effort has been invested in recent years in the development of novel approaches aimed at the specific suppression of unwanted gene expression leading to viral and malignant diseases. The sequencing of the human genome and the elucidation of many molecular pathways that are important in disease have provided unprecedented opportunities for the development of new therapeutics. So-called „antisense“ oligonucleotides, inhibiting gene expression by creation of a helical complex with target mRNA (carrying ”sense” genetic information), represent a perspective approach in chemotherapy. A potential means to improve the efficacy of steric-blocking antisense oligonucleotides is to increase their affinity for a target RNA. The grafting of cationic amino groups to the backbone of the ON is one way to achieve this. Cationic a-ON with phosphoroamidate internucleoside linkages incorporated into ON with a non-natural a-anomeric configuration bound with high affinity to single-stranded DNA and RNA targets [1]. Duplex stabilization is proportional to the number of cationic modifications, with fully cationic ON having particularly high thermal stability. The average stabilization is greatly increased at low ionic strength. Cationic a-ON are well suited as steric-blocking antisense agents as demonstrated in the case of hepatitis C virus [1]. Interestingly, no vectorisation was necessary for the cationic a-ON in the cell culture.
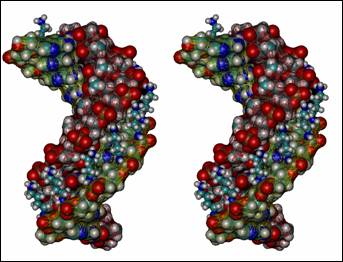
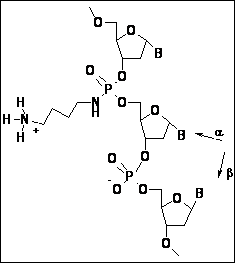
The present work deals with cationic a-ON analogs of nucleic acids. Various side-chain linkers enhancing ability of the a-deoxyadenosine strand to create a double helical complex with the target DNA or RNA strand were tested. Linkers were based on either lysine (butyl-amino phosphoramidates) or arginine (butyl guanidinium phoshoroamidates) side chains (naturally occurring in proteins). Several double helical structures (parallel oriented) consisting of the natural chain (either dT12 or rU12) and cationic a-deoxyadenosine counterpart were used as model systems. Fully solvated molecular dynamics simulations revealed that both types of tethers are able to bridge the minor groove (slightly narrowed in the case of atypical parallel oriented a/b hybrid duplexes) and interact efficiently with phosphate groups in the complementary natural strand. Direct hydrogen bonds between positively charged heads of linkers and negatively charged phosphate groups were established within the MD runs. Average values as well as time-development of distances between groups participating in the aminoalkyl-phosphate hydrogen bonding were analyzed in details. Differences between a-DNA:DNA and a-DNA:RNA systems (as a consequence of various minor groove width) were found.
In
acknowledgments, this work was supported by the Grant of the Ministry of
Education, Youth and Sports of the Czech Republic (project No. MSM 0021620835) and the Grant Agency of Czech
Republic (project No. 203/05/0827 and No. 202/02/D114). Results have been partially obtained using computer
facilities of the Metacentrum of the Czech Universities.
1) T. Michel, C. Martinand-Mari, F. Debart, B. Lebleu, I. Robbins and J. J. Vasseur: „Cationic phosphoroamidate a-oligonucleotides efficiently target single-stranded DNA and RNA and inhibit hepatitis C virus IRES-mediated translation“ Nucleic Acids Research, 2003, Vol. 31, No. 18 5282-5290