MOLECULAR DYNAMICS STUDY OF NOVEL RNA-RNA INTERACTIONS MOTIFS: RNA KISSING COMPLEXES
Kamila Réblová1 ,
Naďa Špačková2, Judit E. Šponer2, Jaroslav Koča1
and Jiří Šponer2,*
1National Center for Biomolecular Research, Masaryk
University, Kotlařská 2, 611 37 Brno, Czech Republic
2National Center for Biomolecular Research, and
Institute of Biophysics, Academy of Sciences of the Czech, Republic,
Královopolská 135, 612 65 Brno, Czech Republic
Explicit solvent molecular dynamics (MD) simulations were carried out for three RNA kissing-loop complexes. The theoretical structure of two base pairs (2 bp) complex of H3 stem-loop of Moloney murine leukemia virus agrees with the NMR structure with modest violations of few NMR restraints comparable to violations present in the NMR structure. In contrast to the NMR structure, however, MD shows relaxed intermolecular G-C base pairs.
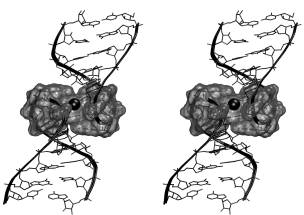
The core region of the kissing complex forms a cation-binding pocket with highly negative electrostatic potential. The pocket shows nanosecond-scale breathing motions coupled with oscillations of the whole molecule. Additional simulations were carried out for 6 bp kissing complexes of the DIS HIV-1 subtypes A and B. The simulated structures agree well with the X-ray data. The subtype B forms a novel four-base stack of bulged-out adenines. Both 6 bp kissing complexes have extended cation-binding pockets in their central parts. While the pocket of subtype A interacts with two hexacoordinated Mg2+ ions and one sodium ion, pocket of subtype B is filled with a string of three delocalized Na+ ions with residency times of individual cations 1-2 ns. The 6 bp complexes show breathing motions of the cation-binding pockets and loop major grooves.