A Phenylnorstatine Inhibitor Binding to HIV-1 Protease: Geometry, Protonation and Subsite-Pocket Interactions Analyzed at Atomic Resolution
Jiri Brynda,* Pavlina Rezacova, Milan Fabry, Magdalena Horejsi, Renata Stouracova, and Juraj Sedlacek
Institute of Molecular Genetics, Academy of Sciences of the
Czech Republic, Flemingovo nám. 2, 16637 Prague6
Milan Soucek, Martin Hradilek, Martin Lepsik, and Jan Konvalinka
Institute of Organic Chemistry and Biochemistry, Academy of
Sciences of the Czech Republic, Flemingovo nám. 2, 16610 Prague 6
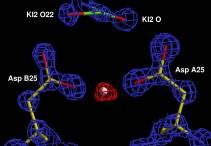
The x-ray structure of a complex
of HIV-1 protease (PR) with a phenylnorstatine inhibitor Z‑Pns‑Phe‑Glu‑Glu‑NH2
has been determined at 1.03 Å, the highest resolution so far
reported for any HIV PR complex. The inhibitor shows subnanomolar Ki values for
both the wild‑type PR and the variant representing one of the most common
mutations linked to resistance development. The structure displays a unique
pattern of hydrogen bonding to the two catalytic aspartate residues. The high
resolution permits to assess the donor/acceptor relations of this hydrogen
bonding, and to indicate a proton shared by the two catalytic residues.
Structural mechanism for the unimpaired inhibition of the protease Val82Ala
mutant is also suggested, based on energy calculations and analyses.