Biomolecular interactions studied by real-time label-free SPR technology
Ivana Nemčovičová
Institute of Virology, Biomedical Research Center, Slovak Academy of Sciences, Bratislava, Slovakia
ivana.nemcovicova@savba.sk
Very little happens in any biological system unless two or more molecules come together to form a stable complex [1]. When molecules interact through specific molecular contacts, all of the principles of thermodynamics, dynamics, and biomolecular structure and recognition come into play. As increasing numbers of new proteins and DNA sequences are entered into databases such as SWISSPROT or GenBank, rapid methods to accurately characterize these biointeractions are needed. One useful model to consider involves a target molecule (T) with a specific binding site (such as a particular region in a protein tertiary structure or a specific sequence of DNA) and a probe molecule (P) that can bind to that site. The simplest binding model corresponds to P + T → C, where C is the resulting complex. Probe molecules can vary from small metabolites or drugs to large transcription complexes, and their interactions with the target range from the highly specific (P binds a single site) to the nonspecific (P binds most sites in the target class, such as related DNA sequences). In interaction processes that are complicated, there can be multiple binding sites, cooperative interactions, and so forth [1-3]. In order to determine the equilibrium and/or kinetic constants for binding, all techniques must factor the concentrations of P and T into the concentrations of free P and T, on the left side of the equation, as well as the concentrations in C, on the right side of the equation. This evaluation can be achieved by various methods, including equilibrium dialysis, spectral measurements, gel shift, calorimetry, DNase I footprinting, and related techniques [1-4]. Many of these methods require labeling of P or T with a fluorescent or radioactive tag.
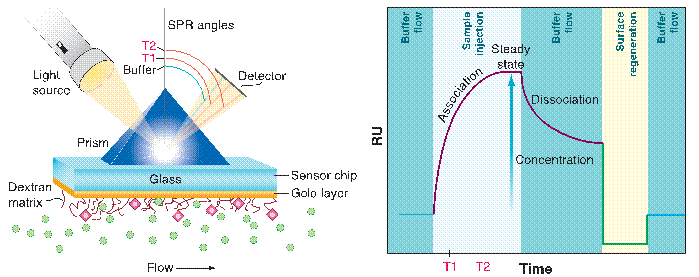
Figure 1: Illustrated SPR. At left, an SPR optical unit and a sensor chip detect the P molecules (green spheres) in the flow solution, which passes by the T (pink diamonds) linked to the dextran matrix. The blue SPR angle defines the position of the reduced-intensity beam. Time points T1 and T2, shown in the schematic sensorgram (right) correspond to the two red SPR angles, which shift as P binds to T over time. As the concentration of bound P increases (arrow), the RU response approaches saturation. The complex dissociates upon reintroduction of the buffer. As shown, the response to the injection solution will fall below the baseline if its refractive index is lower than that of the buffer [Figure adapted from reference 1, Wilson et al.].
A recent development in instruments that investigate biomolecular interactions in label-free mode, is surface plasmon resonance (SPR) detection with a biospecific sensor chip [1-4]. In BIACORE technology, the sensor chip is created by applying a thin layer of gold (~50 nm) to a glass surface (Figure 1, left). In the most common type of sensor chip, carboxymethyl-dextran is linked to the gold to give the interaction layer (~100 nm thick). One of the interacting molecules, either T or P, must be linked to this layer to create the biospecific recognition surface. Because SPR responds to changes in refractive index (Figure 1, right) and, thus, to changes in mass, it is advantageous to attach the molecule with the lowest molecular weight to the surface.
1. W. David Wilson, Science. 295, 2103-5 (2002), and references therein.
2. C. R. Cantor, P. R. Schimmel, Biophysical Chemistry (Freeman, New York, 1980), vol. III.
3. L. G. Fagerstam et al., J. Chromatogr. 597, 397 (1992), and references therein.
4. BIAtechnology Handbook, (BIACORE AB, Uppsala, Sweden, 1998)
IN is Marie Curie Fellow financed by Programme SASPRO, co-funded by European Union and the Slovak Academy of Sciences under the contract No. 0003/01/02. IN acknowledges the contribution of the Slovak Research and Development Agency under the project APVV-14-0839 and the contribution of the Scientific Grant Agency of the Slovak Republic under the grant 2/0103/15.