Sedimentation analysis of proteins using analytical ultracentrifuge: old method with new horizons
O. Vaněk
Department of Biochemistry, Faculty of Science, Charles University, Hlavova 2030, 12840 Prague, Czech Republic
ondrej.vanek@natur.cuni.cz
Sedimentation analysis of macromolecules carried out in analytical ultracentrifuge is a powerful method for the study of proteins, nucleic acids and other polymers and their various complexes. Monitoring sedimentation of macromolecules in the centrifugal field allows their hydrodynamic and thermodynamic characterization in solution, i.e. in native conditions, without interactions with any matrix or surface. This allows direct measurement of molecular weight and sedimentation coefficient of macromolecules, monitoring of sample purity and homogeneity, prediction of size and shape of sedimenting species and, last but not least, study of equilibrium reactions, including determination of their stoichiometry and equilibrium constants.
In the present lecture, we will first focus on the history physical principles of the technique and also on the properties and potential of a modern instrumentation. Two types of experiments performed using analytical ultracentrifuge (i.e. sedimentation velocity and sedimentation equilibrium) will be discussed, together with a brief introduction into sedimentation theory. In the end, sedimentation data analysis will be described and examples of utilization of analytical ultracentrifugation in biomolecule research will be provided.
Combination of new instrumentation and computational software for data analysis has led to major advances in characterization of proteins and their complexes. After temporary silence in the past decades, analytical ultracentrifugation at presence experiences renaissance in proteomic and structural biology research while still being heavily used e.g. for characterization of aggregation of monoclonal antibodies in biopharmaceutical industry.
|
|
|
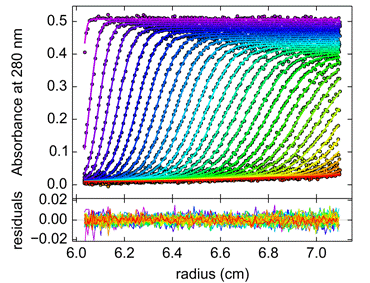
Figure 1. Sedimentation velocity (SV) analysis of recombinant human proliferating cell nuclear antigen (PCNA); fitted data (top) and residuals plot (bottom). |
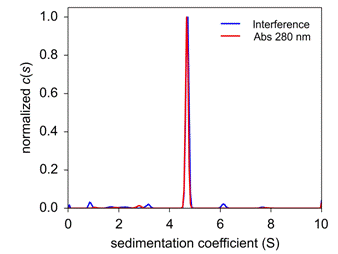
Figure 2. Distribution of sedimenting species for the PCNA SV analysis performed with both absorbance and interference optics showed it is a trimer. |